http://www.biosciencemag.org August 2013 / Vol. 63 No. 8 • BioScience 657
by Jonathan G. Lundgren and Jian J. Duan
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
YOU ARE REQUIRED TO READ THE COPYRIGHT NOTICE AT THIS LINK BEFORE YOU READ THE FOLLOWING WORK, THAT IS AVAILABLE SOLELY FOR PRIVATE STUDY, SCHOLARSHIP OR RESEARCH PURSUANT TO 17 U.S.C. SECTION 107 AND 108. IN THE EVENT THAT THE LIBRARY DETERMINES THAT UNLAWFUL COPYING OF THIS WORK HAS OCCURRED, THE LIBRARY HAS THE RIGHT TO BLOCK THE I.P. ADDRESS AT WHICH THE UNLAWFUL COPYING APPEARED TO HAVE OCCURRED. THANK YOU FOR RESPECTING THE RIGHTS OF COPYRIGHT OWNERS.
The potential hazards posed by RNA interference (RNAi)–based pesticides and genetically modified crops to nontarget organisms include off- target gene silencing, silencing the target gene in unintended organisms, immune stimulation, and saturation of the RNAi machinery. Nontarget organisms will vary in their exposure to small RNAs produced by genetically modified crops, but exposure to insecticidal small RNAs will probably occur at a previously unrealized scale for many. Areas that warrant future work include the persistence of insecticidal small RNAs in the environment, describing crop-based food webs to understand those species that are most exposed, sequencing genomes for species to proactively understand those that may be affected by RNAi, and substantiating that laboratory toxicity testing can accurately predict the field-level effects of this technology. The costs and benefits of pesticidal RNA must be considered relative to current pest management options as pesticidal RNAs move from a theoretical approach to being used as a practical tool.
Keywords: exposure, GM crops, insecticide, nontarget testing, RNAi
Modern pest management has evolved alongside recent developments in crop production practices, and the speed at which new technologies for pest management are advancing challenges our ability to predict and assess the potential ecological risks associated with these technologies. Current insect-resistant genetically modified (GM) crops are well tailored to fit within modern crop production practices, but these technologies face challenges and will need to adapt to accommodate increasing demands on crop production. Additional pest management tools are needed to keep up with future agricultural demands, and RNA interference (RNAi)–based insecticides and GM crops are one response to this impending problem (box 1).
There is a growing interest in using RNAi for insect control, both as a traditionally applied insecticide and within GM plants. RNAi-based GM plants targeting insects have been developed in three independent research programs, although additional GM crops are in development. Baum and colleagues (2007) developed GM corn plants that resisted the western corn rootworm (Diabrotica virgifera; Coleoptera: Chrysomelidae). By reducing translation of vacuolar H+-ATPase subunit A (v-ATPase A) in the pest, the plant increased pest mortality and larval stunting and experienced less root damage as a result. Zha and colleagues (2011) transformed rice plants to suppress the expression of several genes in Nilaparvata lugens (Hemiptera: Delphacidae), a major pest. Although gene expression was suppressed, the insects were not killed by feeding on the GM rice plants. In a different approach to pest management, Mao and colleagues (2011) transformed cotton plants to produce double-stranded RNA (dsRNA) that reduced the expression of the P450 gene CYP6AE14 in cotton bollworms (Helicoverpa armigera; Lepidoptera: Noctuidae). This P450 degrades gossypol, an antiherbivore phytochemical produced by cotton. GM cotton plants experienced less damage than the conventional plants did, and the larvae that were fed the GM cotton had reduced growth but were not killed. These examples illustrate that the creation of RNAi-based GM crops that are lethal to pests or that deleteriously affect interactions of the pests with other organisms (including the crop) is a very real technology that has potential for limiting the impact of pests on crops.
Box 1. What is RNAi?
RNA interference (RNAi) is a posttranscriptional technique for the sequence-selective silencing of genes (Agrawal et al. 2003, Siomi and Siomi 2009). Fragments of small RNAs (small interfering RNAs [siRNA] or microRNAs) bind to messenger RNAs (mRNAs) and promote cleavage by a complex of enzymes, thereby reducing the expression of specific genes. For decades, RNAi was known to occur in plants (as posttranscriptional gene silencing) and fungi (as quelling) but was only first reported in animals (the nematode Caenorhabditis elegans) in 1998 (Agrawal et al. 2003). A cell produces double-stranded RNAs (dsRNAs) or microRNAs that target mRNAs from a specific gene, depending on nucleotide sequence, or dsRNAs are taken into a cell from the exterior environment (environmental RNAi; Huvenne and Smagghe 2010). The dsRNA (generally fewer than 1000 nucleotides [nt] long) is then cleaved into much smaller siRNAs (almost always 21–23 nt long), which are sometimes amplified intracellularly (Siomi and Siomi 2009). It is noteworthy that this amplification has not been widely found in insects (a primary target of RNAi-based GM crops; an exception is embryonic Drosophila melanogaster) or mammals (Agrawal et al. 2003, Dillin 2003). The siRNAs are incorporated into an RNA-induced silencing complex (RISC), where mRNAs are cleaved with an enzyme in the Argonaute family, and their translation is silenced. Silencing in the absence of cleavage may result if the RISC unit simply binds to an mRNA, thereby restricting its translation (Alemán et al. 2007). RNAi is not a way to knock out gene expression, only a way to suppress it, and sometimes only temporarily.
Risk posed by RNAi-based GM crops
There are similarities and differences in the risks associated with insecticidal RNAi relative to those posed by chemical and microbial pesticides and Bt crops, which have pesticidal effects derived from the bacteria Bacillus thuringiensis (Heinemann et al. 2013). Risk is often assessed for pesticides and Bt crops using a tiered approach that relies on a maximum-hazard dose testing regimen targeted at indicator species. Laboratory toxicity assays involve administering nontarget species a maximum-hazard dose (1–20 times the dose) of the known environmental exposure concentration (Harrap 1982, USEPA 2002, 2003). The tests are often focused on six to eight indicator species (such as honeybees, springtails, earthworms, daphnia, predatory beetles or pirate bugs, and parasitoid wasps), which represent different functional guilds (e.g., pollinators, predators, parasitoids, detritivores; USEPA 1994, 1998). A good example of this approach involves Bt crops.
Since the early 1990s, the maximum-hazard dose regimen has been used to characterize the level of risk of several classes of insecticidal Cry proteins produced by the entomopathogen B. thuringiensis that are expressed in Bt crops. To date, this testing regimen has revealed no toxicity of Cry proteins to the selected indicator species (O’Callaghan et al. 2005, Romeis et al. 2006, Duan et al. 2008, 2010). In part, this is because Bt crops have a very narrow and predictable activity spectrum. This specificity is related to the physiological conditions of the insect gut, especially the presence of specific receptor sites on the midgut epithelium (van Frankenhuyzen 2009, Jurat-Fuentes and Jackson 2012). The long historical use of Bt as a microbial pesticide provided crucial baseline information on the mode of action of Bt against susceptible insects, which has helped scientists understand the results of maximum-hazard dose testing involving Bt crops. The effects of Bt toxins on the field abundance of nontarget organisms is often (but not always) predictable using the maximum-hazard dose regimen (Duan et al. 2010), and, arguably, the commercialization of Bt crops has had few if any consistent direct effects on the abundance of nontarget organisms under field conditions (Marvier et al. 2007, Duan et al. 2008, Wolfenbarger et al. 2008, Lundgren et al. 2009, Naranjo 2009, Peterson et al. 2011). For chemical and microbial pesticides and Bt crops, the modes of action are well described, and laboratory nontarget toxicity assays can be focused and optimized on the basis of predictable effects. Although many aspects of the risk assessment of RNAi are similar to those used to assess the risks of other GM crops and pesticides, there is a crucial difference between these technologies that pertains to the mode of action of small RNAs. Small RNAs often have off- target binding elsewhere in a nontarget species’ genome that makes predicting toxic effects and designing maximumhazard dose assays challenging for the wide range of species potentially exposed. This conclusion is in contrast to that of McLean (2011), which was that the maximum-hazard dose paradigm would sufficiently address the risk of RNAi-based technologies. Some potential hazards of small RNAs and exposure pathways are presented in detail below.
The risk posed by RNAi used by plants and other organisms to regulate gene expression, cellular development, and to combat transposon or viral invaders differs from that of insecticidal small RNAs. Evidence suggests that RNAi may have originally evolved within eukaryotes as a way to combat infections from viruses and transposons (Agrawal et al. 2003). GM RNAi-based plants that resist viral phytopathogens are currently commercially available (Mansoor et al. 2006, Auer and Frederick 2009). However, insecticidal RNAi differs from RNAi used in plants to combat viral pathogens in that—to the best of our knowledge—RNAi is not used by plants in the natural world to silence critical gene functions in herbivores. Insecticidal small RNAs are specifically selected or designed to overcome cellular defenses and barriers to small RNAs in order to kill a higher organism. With barriers overcome, genes in higher organisms may be more exposed to insecticidal small RNAs than they are to antiviral small RNAs.
Hazards posed by RNAi to nontarget organisms
Although small interfering RNAs (siRNAs) were originally believed to be extremely specific (Dillin 2003), recent experience with RNAi in functional genomics has revealed that siRNAs often silence unintended genes (Davidson and McCray 2011). Moreover, the process of RNAi can affect organisms in ways that transcend the effects of gene silencing. The hazards of siRNAs within nontargets can be categorized as off-target gene silencing, silencing the target gene in nontarget organisms, immune stimulation, and saturation of the RNAi machinery (this list is adapted from Jackson and Linsley 2010). Knowledge gaps in the genomics and physiologies of highly exposed nontarget organisms currently preclude our ability to assess the activity spectrum of RNAi, determine whether toxicity assays will be sufficient in predicting the risks of RNAi-based crops, and explain how these risks may affect food webs associated with agroecosystems. This last knowledge gap is not unique to RNAi-based technologies.
The specificity of siRNAs for a specific messenger RNA (mRNA) is linked to a certain minimal level of sequence homology. Perfect sequence homology between an mRNA and the dsRNA expectedly results in suppression of the targeted mRNA (Elbashir et al. 2002) but represses the phenotype to varying degrees, depending on the mRNA selected. Substantial sequence divergence between the two molecules does not preclude gene silencing (Saxena et al. 2003). In part, this is because the dsRNA is cleaved into numerous, very short (21–23 nucleotides [nt]) siRNAs that have abundant direct sequence matches throughout the genomes of most organisms. This consistent size of siRNAs optimizes the specificity of the siRNA for the target mRNA relative to the likelihood of off-target binding (Qiu et al. 2005) but does not preclude off-target effects for nontarget organisms (Jackson and Linsley 2010). Quite the contrary, it appears that RNAi operates within cells using a certain level of redundancy among targets (Jackson et al. 2006). One way to reduce potential nontarget effects may be to engineer plants to produce siRNA or microRNA of a known sequence rather than dsRNAs that are subsequently cleaved, but this may reduce the likelihood of silencing the target gene, as well. Recent research has shown that sequence identity in the final 2–8 nt of the 5′ end of the guide strand of siRNA (dubbed the seed region; this corresponds to the 3′ untranscribed region [UTR] of the mRNA) is the only homology necessary for some level of silencing of both target and off-target genes (Jackson et al. 2006, Jackson and Linsley 2010). Once this requisite seed region sequence is matched, additional sequence homology and characteristics can encourage the fidelity of the reaction with the target. But even the most rational dsRNA design does not preclude some level of off- target sequence matching and potential off-target gene suppression in nontarget organisms.
Off-target gene silencing.
One conclusion from the recent advances in functional genomics that has important implications for risk assessment of RNAi-based GM crops is that siRNAs commonly have off-target effects within a targeted cell or organism (Davidson and McCray 2011). The first evidence of this comes through in silico comparisons of sequence homologies between siRNAs and sequences present in the targeted organism. One in silico examination of sequence homologies between siRNA sequences and three transcriptomes from diverse organisms revealed that offtarget effects were observed in as few as 5% and up to 80% of the siRNAs assessed (Qiu et al. 2005). Another study showed that 17% of siRNAs had complete sequence homologies with off-target binding sites in the Drosophila melanogaster genome (Kulkarni et al. 2006). Designing siRNA to reduce off-target binding still produced an average off-target binding rate of 10% or greater (Qiu et al. 2005). Given the small sizes of siRNAs, it is not surprising that off-target binding sites are prevalent within the genomes of all organisms evaluated to date. Although off-target binding would not appear to be a concern in target organisms, off-target binding in nontarget organisms is a real hazard posed by RNAi if the nontargets are sufficiently exposed to the RNAi.
Increasing rates of mRNA and protein suppression are often correlated with increasing rates of off-target binding predicted by in silico searches for sequence homologies between siRNAs and mRNAs, especially when the sequences of the seed region, rather than the complete sequence of the siRNA (Birmingham et al. 2006), are considered. Suppression of mRNA by off-target binding reduces some phenotypes (Saxena et al. 2003), although RNAi effects on off-target protein levels tend to be less studied than mRNA regulation. Federov and colleagues (2006) found that 29% of off- target suppression of mRNAs resulted in reduced viability of transfected cells and that sequence characteristics of the dsRNA affected these viability rates. Off-target binding of siRNAs resulted in reduced protein production in 7 of 30 cases involving a cell culture; this off-target suppression of genes was not accompanied by mRNA cleavage but by binding of the siRNA and the RNA-induced silencing complex (RISC) unit with the targeted mRNA (Alemán et al. 2007). Therefore, considering only mRNA levels may overlook some off-target gene silencing (Saxena et al. 2003, Alemán et al. 2007). These studies indicate that off-target effects of siRNAs used in RNAi are probably more common than was initially believed; these effects could have implications for nontarget effects of GM crops if off-target gene suppression occurs in nontarget organisms and if these organisms are exposed to RNAi to a sufficient degree.
Silencing genes in nontarget organisms.
Most of the work on off-target silencing is related to functional genomics within a single organism, and so the question of how dsRNAs affect target and off-target genes in nontarget organisms has received very little attention. Nevertheless, this is a critical factor in the commercialization of insecticide RNAi. Jackson and Linsley (2010) suggested that off-target silencing appears to be more common within the target organism than in nontarget organisms, but this suggestion was based solely on a comparison between humans and mice. A recent study showed that plant-produced microRNAs constitute 5%–10% of human microRNAs and that these are likely taken in with food (Zhang et al. 2012). The amount of plant microRNA found in rat serum increased when the rats were fed diets containing specific plant microRNAs from rice, even when the rice diet was cooked. One specific plant-produced microRNA examined, miR168, was complementary to mRNAs within rat liver cells and reduced the production of proteins involved in regulating levels of low- density lipoprotein in the rat circulatory system. This work indicates that interspecies nontarget binding of siRNAs and microRNAs taken into an animal through food may occur more often than is commonly thought and may influence gene expression in nontarget organisms that ingest siRNAs within plant tissues. In developing GM maize plants resistant to D. virgifera, Baum and colleagues (2007) also examined the effects of a few of the dsRNAs identified for plant transformation on several other beetle species. They found that the dsRNAs that targeted D. virgifera v-ATPase A and E also reduced survival of Diabrotica undecimpunctata and Leptinotarsa decemlineata significantly, even though these pests shared only 79% and 83%, respectively, sequence homologies in these genes with D. virgifera. Off-target binding of dsRNAs that targeted the v-ATPase A and E genes was not examined in this study. In laboratory feeding assays, Whyard and colleagues (2009) did not find increased mortality when Drosophila spp. ingested dsRNAs designed to suppress a congener’s tubulin gene. These results were echoed when more phylogenetically distant insect taxa ingested dsRNAs aimed at repressing other species’ γ-tubulin or v-ATPase expression, although mRNA knockdown for the latter gene was minimal even for the targeted insect species. Additional research in this area will shed light on the potential nontarget effects of insecticidal dsRNAs and will hopefully address whether a focus on toxicity (the focus of published studies thus far) is sufficient for predicting the nontarget effects of RNAi-based crops under field conditions.
Immune stimulation.
The innate immune systems of higher organisms rely on pattern recognition proteins and other factors to identify potentially pathogenic invaders, and these defenses recognize and eliminate dsRNAs that are potential pathogens. Recently, it was found that the injection of small fragments (fewer than 30 nt) of RNA could stimulate an immune reaction in mammals (Robbins et al. 2009). In this group, some Toll-like receptors recognize and respond to the sequence, length, and structure of siRNAs. This has been studied most intensively in mammals, and in mice, the immunostimulation by RNAi led to reduced lymphocytes and platelet cells, largely correlated with cytokine response to the siRNA (Judge et al. 2005). Although there are some similarities in the innate immune response of insects and mammals (Lundgren and Jurat-Fuentes 2012), it is unclear how the immune systems of other organisms will react to an influx of small RNAs. Nor is it known how this immunostimulation will affect the fitness of nontarget organisms. Indeed, the risk of immunostimulation by dsRNAs may be one reason for which the enzyme RNA-dependent RNA polymerase (RdRP), which is responsible for amplifying the abundance of siRNAs in some organisms, has yet to be found in mammals and insects (Agrawal et al. 2003, Dillin 2003). Although slight changes in nucleotide sequence can mitigate many immunostimulatory effects in a given organism (Jackson and Linsley 2010), substantial research will be required if we are to determine the effects of RNAi inputs on the immune responses of members of entire biological communities associated with agroecosystems.
Saturation of the RNAi machinery.
High levels of exogenous siRNAs can saturate a cell’s RNAi machinery and thereby reduce the efficiency at which a cell regulates endogenous gene expression (Agrawal et al. 2003, Dillin 2003). Essentially, there is a limited number of RISCs present within a cell, and if the augmented siRNAs saturate these complexes, the health and performance of the cell may be compromised (Kahn et al. 2009). Jackson and Lindley (2010) found evidence that small RNAs could have “global effects on the expression of genes predicted to be under the control of endogenous microRNAs” (p. 64). This process of saturation is better documented with small hairpin RNA (a type of siRNA that targets a specific place on the mRNA), although it is known from siRNA as well (Jackson and Linsley 2010). The degree to which a nontarget species is exposed to a specific pesticidal small RNA needs to be considered when saturation potentials are discussed. Suffice it to say that it is unclear how dsRNAs produced by plants could affect the RNAi machinery used by both target and nontarget organisms and whether there will be sufficient small RNA produced by GM plants to saturate an organism’s cellular machinery.
Exposure to RNAi-based GM crops
Even the most toxic pesticides pose no risk to nontarget organisms if the organisms are not exposed physically or physiologically to these toxins in the environment. There is a substantial number of nontarget species that will be exposed to RNAi-based crops if planting is widespread, but the exposure level for each of the myriad species remains difficult to predict. This exposure includes physical exposure to the toxin (i.e., being in the right place at the right time), but it also involves the organism’s having the correct physiological characteristics (e.g., receptor sites, genetic sequences, cellular machinery) to allow the toxin to work if it is physically exposed.
Physical exposure to insecticidal RNAi.
A large number of nontarget organisms will likely be physically exposed to insecticidal small RNAs if RNAi-based crops and RNAibased insecticidal sprays are commercialized and widely used, but this physical exposure would be similar to that experienced by current GM crops and systemic chemical and endophytic microbial pesticides. Physical exposure is also constrained to those organisms that consume the toxin. However, much of the non- and off-target work on RNAi has been conducted in a Petri dish, so understanding the physical exposure to small RNAs at this greatly amplified scale is important. In 2011, nearly 10% (735,000 square kilometers) of the land surface in the continental United States was planted with three plant species (corn, soybean, and cotton), each of which is currently genetically modified to facilitate pest management (insects or weeds; see figure 1; http://www.nass.usda.gov/index.asp). Although biological inventories of these agroecosystems have been pursued by biologists for more than 100 years, we still have a poor resolution of the large number of eukaryotic species (most of which possess the machinery for and use RNAi) that reside within these habitats. Nevertheless, the numbers available suggest that each community has several hundred species. For example, hundreds of arthropod species reside within cornfields, and these dynamic communities change over the season and vary by region (Bhatti et al. 2005, Dively 2005, Lundgren and Fergen 2010). Add to this inventory fungi, noninsect animals, and noncrop plants that use RNAi, and the list of species in this habitat expands substantially. The roles that most of these species play in healthy ecosystem functioning are entirely unknown. Considering the current footprint of GM crops on the terrestrial landscape and the number of species residing in those crop habitats, a significant number of species will be exposed to RNAi-based crops if this technology becomes adopted at a level comparable to that of current GM crops.
Physiological exposure to insecticidal RNAi.
Many nontarget organisms have characteristics that will allow them to be physiologically exposed to insecticidal small RNAs; this differs from narrow spectrum insecticides such as Bt. If possessing the correct 23-nt gene sequence were all that dictated the physiological exposure to insecticidal RNAs, nearly all physically exposed organisms would be considered physiologically exposed (these short nucleotide sequences are randomly present in many genomes); clearly, this is not the case. Higher organisms also present numerous barriers (e.g., physiological gut conditions, specificity of RNAi enzymatic machinery) to restrict unwanted gene silencing by ingested small RNAs. Understanding the physiological basis of RNAi reveals several levels of physiological characteristics that will winnow the number of nontarget species ultimately exposed to unintentional gene silencing by insecticidal RNAs.
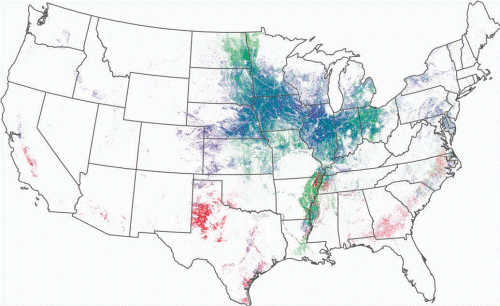
Figure 1. In 2011, approximately 26% of the land surface (558,000 square kilometers) in 12 Midwestern states (Illinois, Indiana, Iowa, Kansas, Michigan, Minnesota, Missouri, Nebraska, North Dakota, Ohio, South Dakota, and Wisconsin) was planted almost exclusively with two plant species: corn and soybean, both of which are genetically modified for weed and insect pest management. In the map, red marks cotton fields, green is corn, and blue is soybean. The data, from 2011, were generated using the US Department of Agriculture National Agriculture Statistics Service’s Cropland Data Layer Program data set.
Organisms ingest small RNAs with every meal, and this obviously does not appear to silence gene functions. Environmental and physiological conditions in the gut probably destroy many small RNAs taken in with food (Wang J et al. 2010, O’Neill et al. 2011). Those small RNAs that survive must be adapted to function within an organism. Different organisms have slight deviations in the receptors that allow transmembrane movement of dsRNAs (SID1, SID2) and in enzymes that direct RNAi (e.g., Dicer, Argonaute, RdRP, RNA and DNA helicases; Agrawal et al. 2003, Siomi and Siomi 2009). These enzymes often have similar or identical functional domains, and knowledge gaps make it unclear how dsRNAs that target a pest will function within the RNAi pathways of other organisms (especially phylogenetically divergent ones). What is clear is that insecticidal small RNAs are selected or designed to suppress genes within arthropods after ingestion and, therefore, possess mechanisms that allow them to overcome the restrictions that prohibit the function of the myriad other small RNAs ingested with every meal. We hypothesize that nontarget taxa that are phylogenetically close to the targeted pest will be most likely to have similar RNAi pathways and suggest that these taxa are most likely to be affected by RNAi; additional information on other species is necessary to substantiate this assumption.
In a sense, mRNAs are in an arms race with RNAi, and the nucleotide sequences of both drive which genes might be affected by a particular RNAi. Genes whose regulation is tied to RNAi tend to have longer 3′ UTRs with more potential seed regions that facilitate binding of siRNAs and microRNAs; those mRNAs that are not targeted by RNAi have shorter 3′ UTRs (Jackson et al. 2006). These untargeted mRNAs can also regulate their expression so that coexpression with mRNA targets is avoided (Qiu et al. 2005, Jackson et al. 2006). The structure of the siRNA and mRNA in question also has important effects on the outcome of off-target RNAi. Modifying the second position of the seed region of an siRNA by substituting it with O-methyl ribosyl can reduce but not eliminate off-target binding within a target organism (at least in cell lines; Jackson and Linsley 2010). The concentration of the small RNA and the level of gene expression dictate which genes will be suppressed in specific tissues and at what level (Elbashir et al. 2002, Jackson and Linsley 2010). Much of the focus in off-target studies also centers on the sequences of the siRNA and the corresponding region of the mRNA, but sequences in the mRNA that surround the homologous region also affect whether a specific mRNA will be bound to a RISC. Although the knowledge gained from each study improves our ability to predict the outcome of RNAi, we still do not fully understand all of the reasons that RNAi functions only some of the time (Jackson and Linsley 2010). Suffice it to say that knowledge gaps reduce our ability to predict when the fitness and performance of nontarget organisms will be affected, even when in silico comparisons between siRNAs and nontarget genomes suggest that binding is likely.
Knowledge gaps in nontarget effects of RNAi-based crops and some potential solutions
Not all of the hundreds of species living in agroecosystems will be equally exposed to the pesticidal RNAs or will have measurable levels of hazard. Before we are able to weigh the risks to nontarget species against the benefits gained from protecting crops from herbivory, there are a number of knowledge gaps regarding the nontarget effects of RNAi- based insecticides that merit further study, not all of which are unique to RNAi-based technologies.
The persistence of dsRNAs and siRNAs in the environment and the movement of these molecules throughout the landscape are largely unknown, but their persistence will affect the degree to which nontarget organisms are exposed (Auer and Frederick 2009, McLean 2011). Methods have been developed for detecting the degradation of nucleic acids in various soils (Levy-Booth et al. 2007), and it is feasible that some of the technologies developed for studying DNA degradation in the soil could be adapted to small RNAs (Wang Y et al. 2009). Key considerations in RNA degradation rates include the biological, chemical, and physical aspects of the soil (Levy-Booth et al. 2007, Pietramellara et al. 2009). Nucleic acids—DNA has been studied the most in this regard—in the soil can persist by binding to humic substances and minerals, can be degraded by microbes or extracellular deoxyribonucleases, or can be incorporated into microbial genomes (Levy-Booth et al. 2007, Pietramellara et al. 2009). DNA from crop plants can persist in the soil for as little as 7 days but for as long as many years (Levy-Booth et al. 2007, Nielsen et al. 2007). For good evolutionary reasons, RNA seems to degrade more quickly than DNA in the soil, but structural aspects of the RNA molecule (e.g., hairpins) and the degradation rates of plant tissues harboring the RNAi may facilitate the persistence of these molecules in the environment. If transgenes or small RNA products are taken into microbial genomes, this will have implications for which species are trophically exposed to plant-derived RNAi. Environmental persistence of insecticidal toxins—be they chemical, microbial, or nucleic acids—depends on various aspects of the soil, environment, and biological community within a habitat. Understanding the relative degradation rates of these myriad compounds will be important in assessing the costs and benefits of RNAi-based technologies relative to those of other insecticides.
Poor resolution of crop-based food webs prohibits knowing which species will be exposed to crop-expressed dsRNAs. The primary route of exposure to pesticidal RNAi is trophic in nature, as it is in current insecticidal GM crops. Recent advances in tracking foods through food webs offer a good opportunity for quantitatively and empirically narrowing the list of species that may be exposed to RNAi technology. Gut content analysis (searching for a food-specific marker within the stomachs or feces of field-collected animals; e.g., Weber and Lundgren 2009) can identify which species directly consume a crop species (or a crop’s DNA) in the field (Harwood et al. 2005, Zwahlen and Andow 2005). The reliability of these linkages is based on the technique’s specificity for a food-associated marker. Although there are limitations to the technique (Weber and Lundgren 2009), polymerase chain reaction–based gut analysis has rapidly become the tool of choice for many within this field because of its cost effectiveness and the specificity possible with primer sequences (Weber and Lundgren 2009). The relative frequencies of consumption of the GM crop can be used to develop a focused list of species that can be further examined under various hazard scenarios.
The activity spectrum of RNAi is ultimately sequence based, and genomic information on most species is sparse (Auer and Frederick 2009). Recent advances in next- generation sequencing technologies (Metzker 2010) will permit studies that proactively identify targets for pesticidal RNAi on larger numbers of species within an exposed community. The costs of sequencing entire genomes are low enough that it is now feasible to sequence multiple species from members of a biological community that may be exposed to the small RNAs (determined on the basis of food-web analysis). Coordinated efforts to sequence entire genomes of 10,000 vertebrate species (Haussler 2009) and 5000 insect species (Robinson et al. 2011) are currently under way. The sequence homologies between small RNAs expressed by a GM plant and key species in a crop habitat could then be compared in silico using modern algorithms designed for searching for RNAi off-target homologies (McLean 2011).
It is unclear whether traditional toxicity assays are appropriately attuned to determine the effects of RNAi on the fitness and performance of nontarget organisms. The physiological effects of RNAi on nontarget organisms are difficult to predict without some knowledge of which genes are at risk of being silenced by specific small RNAs. Given the mechanism of RNAi (gene suppression), it is possible that the nontarget effects experienced would be sublethal; it is unlikely that these effects would be measurable by looking at survival over time in the laboratory. Some evaluations of the nontarget effects of Bt crops and traditional insecticides advocate the use of the intrinsic rate of population growth, which can be measured in the laboratory and which integrates survival, development time, reproduction, and sex ratio to generate a clearer picture of how a plant-incorporated toxin affects a nontarget species (Bøhn et al. 2010, Li and Romeis 2010). Assessments that provide a comprehensive view of various life-history traits may be justified, given the mode of action posed by RNAi.
Are laboratory assays sufficient for assessing the effects of RNAi? As was discussed at length earlier, RNAi suppresses phenotypes by prohibiting the translation of mRNAs, and it may be challenging to predict which target or off-target genes will be suppressed in nontarget organisms solely on the basis of sequence homologies. Since mRNAs are transcribed as they are needed by the organism, it is important to recognize that the environment plays a significant role in gene expression and, therefore, in which genes will be exposed to the inhibitory small RNAs (Smith and Kruglyak 2008). As a result, conducting hazard assays under controlled conditions within a laboratory may change or reduce the expression of genes that are potential off-targets in nontarget organisms. If gene expression is reduced or altered under laboratory conditions, it may be appropriate to conduct field-based assessments of RNAi-based, insectresistant crops against nontarget organisms, regardless of the outcome of laboratory-based hazard testing.
Conclusions
The rapid development of RNAi applications has challenged scientists to identify and fill key knowledge gaps that underlie the environmental implications of large-scale, pesticidal RNAi-based crops. Much of what we know regarding RNAi comes from the field of functional genomics and the development of gene therapies within individual organisms or even within a specific tissue. How specific small RNAs affect diverse nontarget communities merits further attention, especially in light of the frequent off-target effects of siRNAs within a single target organism. New technologies involved in food web analysis and next-generation sequencing are likely to facilitate the development of risk-assessment frameworks for RNAi-based crops, particularly by honing the relative exposure levels experienced by members of the nontarget community. Because RNAi effects are sequence-based, proactive identification of species with sequences homologous with putative small RNAs for use in pest control could expedite the selection of small RNAs that balance the maximum effects on the target pests with the minimal effects on nontarget organisms. For example, if an organism does not have the genetic sequences that small RNAs can affect, even maximum exposure doses will not result in hazard. Therefore, targeting genes for pest management that are inherently tied to a single species’ biology (e.g., detoxification pathways, developmental regulatory hormones, or mate- finding signals) may reduce the likelihood of silencing a target gene in a nontarget organism. The flexibility, adaptability, and demonstrated effectiveness of RNAi technology indicate that it will have an important place in the future of pest management, but these benefits should be viewed in light of the relative environmental risks that the technology poses.
Acknowledgments
We thank Tatyana Rand of the US Department of Agriculture Agricultural Research Service (USDA-ARS) for creating the distribution map of genetically modified crops. Jimmy Becnel, Keith Hopper, and Don Weber from USDA-ARS; John Turner of the USDA Animal and Plant Health Inspection Service; Robert Wiedenmann of the University of Arkansas; and Chris Wozniak of the US Environmental Protection Agency provided helpful comments on earlier drafts of the manuscript. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
References cited
Agrawal N, Dasaradhi PVN, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. 2003. RNA interference: Biology, mechanism, and applications. Microbiology and Molecular Biology Reviews 67: 657–685.
Alemán LM, Doench J, Sharp PA. 2007. Comparison of siRNA-induced off- target RNA and protein effects. RNA 13: 385–395.
Auer C, Frederick R. 2009. Crop improvement using small RNAs: Applications and predictive ecological risk assessments. Trends in Biotechnology 27: 644–651.
Baum JA, et al. 2007. Control of coleopteran insect pests through RNA interference. Nature Biotechnology 25: 1322–1326.
Bhatti MA, Duan J[J], Head GP, Jiang C, McKee MJ, Nickson TE, Pilcher CL, Pilcher CD. 2005. Field evaluation of the impact of corn rootworm (Coleoptera: Chrysomelidae)–protected Bt corn on foliagedwelling arthropods. Environmental Entomology 34: 1336–1345.
Birmingham A, et al. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nature Methods 3: 199–204.
Bøhn T, Traavik T, Primicerio R. 2010. Demographic responses of Daphnia magna fed transgenic Bt-maize. Ecotoxicology 19: 419–430.
Davidson BL, McCray PB. 2011. Current prospects for RNA interference– based therapies. Nature Reviews Genetics 12: 329–340.
Dillin A. 2003. The specifics of small interfering RNA specificity. Proceedings of the National Academy of Sciences 100: 6289–6291.
Dively GP. 2005. Impact of transgenic VIP3a × Cry1Ab lepidopteran-resistant field corn on the nontarget arthropod community. Environmental Entomology 34: 1267–1291.
Duan JJ, Marvier M, Huesing J, Dively G[P], Huang ZY. 2008. A metaanalysis of effects of Bt crops on honey bees (Hymenoptera: Apidae). PLOS ONE 3 (art. e1415).
Duan JJ, Lundgren JG, Naranjo S[E], Marvier M. 2010. Extrapolating non-target risk of Bt crops from laboratory to field. Biology Letters 6: 74–77.
Elbashir SM, Harborth J, Weber K, Tuschl T. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26: 199–213.
Federov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A. 2006. Off-target effects by siRNA can induce toxic phenotype. RNA 12: 1188–1196.
Harrap KA. 1982. Assessment of the human and ecological hazards of microbial insecticides. Parasitology 84: 269.
Harwood JD, Wallin WG, Obrycki JJ. 2005. Uptake of Bt-endotoxins by non-target herbivores and higher order arthropod predators: Molecular evidence from a transgenic corn agroecosystem. Molecular Ecology 14: 2815–2823.
Haussler D, et al. 2009. Genome 10K: A proposal to obtain whole-genome sequence for 10,000 vertebrate species. Journal of Heredity 100: 659–674.
Heinemann JA, Agapito-Tenfen SZ, Carman JA. 2013. A comparative evaluation of the regulation of GM crops or products containing dsRNA and suggested improvements to risk assessments. Environment International 55: 43–55.
Huvenne H, Smagghe G. 2010. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. Journal of Insect Physiology 56: 227–235.
Jackson AL, Linsley PS. 2010. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature Reviews Drug Discovery 9: 57–67.
Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12: 1179–1187.
Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. 2005. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature Biotechnology 23: 457–462.
Jurat-Fuentes JL, Jackson TA. 2012. Bacterial entomopathogens. Pages 265–350 in Vega FE, Kaya HK, eds. Insect Pathology, 2nd ed. Academic Press.
Kahn AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. 2009. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nature Biotechnology 27: 549–555.
Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. 2006. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nature Methods 3: 833–838.
Levy-Booth DJ, Campbell RG, Gulden RH, Hart MM, Powell JR, Klironomos JN, Pauls KP, Swanton CJ, Trevors JT, Dunfield KE. 2007. Cycling of extracellular DNA in the soil environment. Soil Biology and Biochemistry 39: 2977–2991.
Li Y, Romeis J. 2010. Bt maize expressing Cry3Bb1 does not harm the spider mite, Tetranychus urticae, or its ladybird beetle predator, Stethorus punctillum. Biological Control 53: 337–344.
Lundgren JG, Fergen JK. 2010. The effects of a winter cover crop on Diabrotica virgifera (Coleoptera: Chrysomelidae) populations and beneficial arthropod communities in no-till maize. Environmental Entomology 39: 1816–1828.
Lundgren JG, Jurat-Fuentes JL. 2012. Physiology and ecology of host defense against microbial invaders. Pages 461–480 in Vega FE, Kaya HK, eds. Insect Pathology, 2nd ed. Academic Press.
Lundgren JG, Gassmann AJ, Bernal J, Duan J[J], Ruberson J. 2009. Ecological compatibility of GM crops and biological control. Crop Protection 28: 1017–1030.
Mansoor S, Amin I, Hussain M, Zafar Y, Briddon RW. 2006. Engineering novel traits in plants through RNA interference. Trends in Plant Science 11: 559–565.
Mao Y-B, Tao X-Y, Xue X-Y, Wang L-J, Chen X-Y. 2011. Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Research 20: 665–673.
Marvier M, McCreedy C, Regetz J, Kareiva P. 2007. A meta-analysis of effects of Bt cotton and maize on nontarget invertebrates. Science 316: 1475–1477.
McLean MA, ed. 2011. Problem Formulation for the Environmental Risk Assessment of RNAi Plants, Conference Proceedings (June 1–3, 2011). Center for Environmental Risk Assessment, International Life Sciences Institute Research Foundation.
Metzker ML. 2010. Sequencing technologies: The next generation. Nature Reviews Genetics 11: 31–46.
Naranjo SE. 2009. Impacts of Bt crops on non-target organisms and insecticide use patterns. CAB Reviews: Perspectives in Agriculture, Veterinary Science, Nutrition, and Natural Resources 4: 1–23.
Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. 2007. Release and persistence of extracellular DNA in the environment. Environmental Biosafety Research 6: 37–53.
O’Callaghan M, Glare TR, Burgess EPJ, Malone LA. 2005. Effects of plants genetically modified for insect resistance on nontarget organisms. Annual Review of Entomology 50: 271–292.
O’Neill MJ, Bourre L, Melgar S, O’Driscoll CM. 2011. Intestinal delivery of non-viral gene therapeutics: Physiological barriers and preclinical models. Drug Discovery Today 16: 203–218.
Peterson JA, Lundgren JG, Harwood JD. 2011. Interactions of transgenic Bacillus thuringiensis insecticidal crops with spiders (Araneae). Journal of Arachnology 39: 1–21.
Pietramellara G, Ascher J, Borgogni F, Ceccherini MT, Guerri G, Nannipieri P. 2009. Extracellular DNA in soil and sediment: Fate and ecological relevance. Biology and Fertility of Soils 45: 219–235.
Qiu S, Adema CM, Lane T. 2005. A computational study of off-target effects of RNA interference. Nucleic Acids Research 33: 1834–1847.
Robbins M, Judge A, MacLachlan I. 2009. siRNA and innate immunity. Oligonucleotides 19: 89–102.
Robinson GE, Hackett KJ, Purcell-Miramontes M, Brown SJ, Evans JD, Goldsmith MR, Lawson D, Okamuro J, Robertson HM, Schneider DJ. 2011. Creating a buzz about insect genomes. Science 331: 1386.
Romeis J, Meissle M, Bigler F. 2006. Transgenic crops expressing Bacillus thuringiensis toxins and biological control. Nature Biotechnology 24: 63–71.
Saxena S, Jónsson ZO, Dutta A. 2003. Small RNAs with imperfect match to endogenous mRNA repress translation. Journal of Biological Chemistry 278: 44312–44319.
Siomi H, Siomi MC. 2009. On the road to reading the RNA-interference code. Nature 457: 396–404.
Smith EN, Kruglyak L. 2008. Gene–environment interaction in yeast gene expression. PLOS Biology 6 (art. e83).
[USEPA] US Environmental Protection Agency. 1994. Proposed policy: Plant-pesticides subject to the federal insecticide fungicide, and rodenticide act and the Federal Food, Drug, and Cosmetic Act. Federal Register 59: 60496–60518.
———. 1998. Guidelines for Ecological Risk Assessment. USEPA.
———. 2002. Guidance on Cumulative Risk Assessment of Pesticide Chemicals That Have a Common Mechanism of Toxicity. Office of Pesticide Programs, USEPA. (14 May 2013; http://www.epa.gov/oppfead1/trac/ science/cumulative_guidance.pdf )
———. 2003. Biopesticides Registration Action Document: Event MON 863 Bacillus thuringiensis Cry3Bb1 corn. Office of Pesticide Programs, USEPA. Van Frankenhuyzen K. 2009. Insecticidal activity of B. thuringiensis crystal proteins. Journal of Invertebrate Pathology 101: 1–16.
Wang J, Lu Z, Wientjes MG, Au JL-S. 2010. Delivery of siRNA therapeutics: Barriers and carriers. AAPS Journal 12: 492–503.
Wang Y, Morimoto S, Ogawa N, Oomori T, Fujii T. 2009. An improved method to extract RNA from soil with efficient removal of humic acids. Journal of Applied Microbiology 107: 1168–1177.
Weber DC, Lundgren JG. 2009. Assessing the trophic ecology of the Coccinellidae: Their roles as predators and prey. Biological Control 51: 199–214.
Whyard S, Singh AD, Wong S. 2009. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochemistry and Molecular Biology 39: 824–832.
Wolfenbarger LL, Naranjo SE, Lundgren JG, Bitzer RJ, Watrud LS. 2008. Bt crop effects on functional guilds of non-target arthropods: A metaanalysis. PLOS ONE 3 (art. e2118).
Zha W, Peng X, Chen R, Du B, Zhu L, He G. 2011. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLOS ONE 6 (art. e20504).
Zhang L, et al. 2012. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Research 22: 107–126.
Zwahlen C, Andow DA. 2005. Field evidence for the exposure of ground beetles to Cry1Ab from transgenic corn. Environmental Biosafety Research 4: 113–117.
Jonathan G. Lundgren (jonathan.lundgren@ars.usda.gov) and Jian J. Duan are research entomologists with the US Department of Agriculture Agricultural Research Service, in Washington, DC. Their expertise is in the ecological risk of pest management technologies and developing biology-based management for insects and weeds.
_______________
Notes:
BioScience 63: 657–665. ISSN 0006-3568, electronic ISSN 1525-3244. © 2013 by American Institute of Biological Sciences. All rights reserved. Request permission to photocopy or reproduce article content at the University of California Press’s Rights and Permissions Web site at http://www.ucpressjournals.com/ reprintinfo.asp. doi:10.1525/bio.2013.63.8.8
