Part 3 of 3
Comments37
Kurt Thomas
JULY 15, 2020 AT 9:50 PM REPLY
Very convincing arguments for the provenance, thank you for researching and sharing.
Lifeguard Larry
JULY 16, 2020 AT 11:01 AM REPLY
If lungs are such great places for the virus to better adapt to human hosts what chance do we have against it now that so many millions of lungs are infected?
M
JULY 16, 2020 AT 7:50 PM REPLY
What is the source of the translation? How did you become aware of the Master’s Thesis?
Jonathan Latham
JULY 17, 2020 AT 12:35 AM REPLY
We hired a Chinese speaker to translate the Chinese Master’s thesis for us quickly, but clearly enough so we could understand the diagnosis and treatment of the miners’ illnesses. The translator was not a doctor, so a doctor kindly checked the English translation to improve clarity/accuracy of the medical terms. We became aware of the Chinese Master’s thesis and later the Chinese PhD thesis (that both reported on the mine incident) after reading the two extremely informative preprints — Rahalkar and Bahulikar 2020 and Segreto and Deigin 2020 — both referenced in the our article. Both are well worth reading.
Charlie Knoll
JULY 16, 2020 AT 9:50 PM REPLY
Your assumption “…BtCoV/4991 and RaTG13 are both from the same bat faecal sample and the same mine. They are thus sequences from the same virus” is suspect at best. BtCoV/4991 only has 373 base pairs. How can you be sure that there were no significant mutations between the unpublished BtCov/4991 portion and RaTG13?
Jonathan Latham
JULY 16, 2020 AT 9:53 PM REPLY
The sequences of both are publicly available so it’s not an assumption. They are identical.
Timothy Jones
JULY 16, 2020 AT 10:38 PM REPLY
excerpt
“As we noted in our earlier article, the most important of the questions surrounding the origins of SARS-CoV-2 could potentially be resolved by a simple examination of the complete lab notebooks and biosafety records of relevant researchers at the WIV.”
Why haven’t these people reconciled the issue with the Bat Woman?
https://www.scientificamerican.com/arti ... onavirus1/
excerpt
“Back in Wuhan, where the lockdown was finally lifted on April 8, China’s bat woman is not in a celebratory mood. She is distressed because stories from the Internet and major media have repeated a tenuous suggestion that SARS-CoV-2 accidentally leaked from her lab—despite the fact that its genetic sequence does not match any her lab had previously studied. Other scientists are quick to dismiss the allegation. “Shi leads a world-class lab of the highest standards,” Daszak says.”
Either she’s lying or the virus is natural.
Charlie Knoll
JULY 16, 2020 AT 11:17 PM REPLY
Ah, sorry I didn’t realize they had published the entire BtCoV/4991 sequence.
Jonathan Latham
JULY 17, 2020 AT 12:39 AM REPLY
Your question is not silly though. Sequencing can have errors, for example. But even if it was a tiny bit different most people would still say it was the same virus on the grounds that its not different enough to be a new strain or species.
Allison Wilson
JULY 17, 2020 AT 12:21 AM REPLY
We hired a Chinese speaker to translate the Chinese Master’s thesis for us quickly, but clearly enough so we could understand the diagnosis and treatment of the miners’ illnesses. The translator was not a doctor, so a doctor kindly checked the English translation to improve clarity/accuracy of the medical terms. We became aware of the Chinese Master’s thesis and later the Chinese PhD thesis (that both reported on the mine incident) after reading the two extremely informative preprints — Rahalkar and Bahulikar 2020 and Segreto and Deigin 2020 — both referenced in the our article. Both are well worth reading.
Dave
JULY 17, 2020 AT 1:05 AM REPLY
Preprint by Xiaoxu Sean Lin and Shihong Chen “Major concerns on the identification of bat coronavirus strain RaTG13 and quality of related Nature paper” doi:10.20944/preprints202006.0044.v1
Jonathan Latham
JULY 17, 2020 AT 1:33 AM REPLY
Excellent question. We agree that questions can be asked about RaTG13. The main one for us is that it is not an infectious clone. I.e. it has not fulfilled Koch’s postulates of being able to infect a bat, cause disease, and fulfill its lifecycle. Therefore it may be a mutant or defective in some way.
M
JULY 17, 2020 AT 2:23 AM REPLY
” Rahalkar and Bahulikar 2020 and Segreto and Deigin 2020″
Have you contacted the authors?
Crocodilite
JULY 17, 2020 AT 6:26 AM REPLY
(Looks like the comments I left yesterday didn’t make it for some reason–perhaps you misunderstood my enthusiasm. I’ll rephrase, be less “colorful,” and try again.)
I think this is an amazingly well-researched and well-thought out coherent piece! Thank you!
(1) About this whole miner saga, this is the first time I’ve heard about the existence of the human tissue and blood samples! If proved true, I think this is incredibly significant!
Do you have witnesses and/or paper trails that could testify to the existence of these samples?
If true, the degree of deception involved in hiding this fact seems mind-boggling to me and this culpability should be highlighted.
In all their public utterance, Shi and company have only cryptically mentioned a tiny sample of bat poop. And have indirectly claimed that the sample is in fact used up, so all traces of anything are gone!
At no place was there ever mention of a big stash of human tissue samples that are, at least according to this piece, likely heavily laden with the virus(es) in question!
The whole legend surrounding the “batwoman” mystique would have been almost an entire fraud, since the most important information was in the abundant and readily available human samples that were shipped to her labs with industrial efficiency, not in the tiny, potentially heavily contaminated poop bits that needed to be painstakingly coaxed out of stinky dark caves!
Exactly what has happened to these samples, I hope, will at some point, become a focal point of inquiry!
Exactly which strain(s) of the virus are in these samples is an incredibly intriguing question. To what extent, if at all, are they different from today’s SARS-CoV-2 or its bat ancestors? Does it include the all-important furin site?
If the samples were destroyed, who did it? Why?
(2) Just WHY?!
The all-important question of why, why did Shi and company hide this entire saga and the existence of the samples, for all these years, the all-important question of *motive*.
I understand it’s kind of beyond the scope of this scientific piece so it was probably prudent for the authors not to go there. But it’s an obvious question at least a reader should ask!
BSL4 is a means; it’s not an end. Waiting for BSL4 is waiting for a means to do what?
The work required for publication, if indeed deemed to be too risky to be carried out in China at the time, could easily be done at any number of overseas facilities, with the aid of Shi’s many international collaborators. Even more dangerous viruses had been studied safely and surely the means existed.
Shi’s entire career is built on the premise that another bat virus after Sars could one day jump from bats to human. She’s been jumping up and down telling the whole world about it. The miner saga is the holy grail! To sit on this for half a decade and keeping quiet about it is not in her character. Sitting on such a dangerous time bomb and being quiet about it is not the best way to warn the world about its impending doom.
And after the miner incident in 2013, she continued splicing experiments, creating worse and worse hybrids. The justification for the splicing experiments was that, since they couldn’t find actual natural cases of bat viruses jumping into humans, they could benefit from artificially creating them so they could study them. The miner incident showed that such jumps did in fact occur in nature and it was indeed as bad as it could get and they had all the samples they needed. That should have removed the excuses for continued splicing experiments.
In summary, I fail to see what legimate work they could possibly have intended that justified waiting for so long and so patiently to perform!
(3) “RaTG13” is a term whose meaning is no longer clear.
It could have been from the bat poop, which, critically, could be very different from what’s in the miner samples, especially in light of the main thesis of this piece, that there had been significant adaptation inside the miner lungs.
It could have come from the miners’ samples.
It could be a hodge-podge of several things in the bat poop.
And, of course, it could even have been faked!
Considering a strong condemnation has been brought against the conduct of Shi and company, the sole source of the dubious information concerning RaTG13, we can no longer confidently invest any meaning or trust in even the existence of RaTG13, let alone the finer details of its essential characteristics.
Reading this piece on its first day of publication has been a privilege and an eye-opener! Thank you!
Philip Ward
JULY 17, 2020 AT 8:48 AM REPLY
One question that immediately comes to mind is that if these patients in 2012 had CoViD-19 then why did this not lead at least to wider local outbreaks – for example in the places where they were treated or amongst their families or the people who transported them to doctors and hospitals. Is there any information about that available?
Lance
JULY 17, 2020 AT 7:14 PM REPLY
Brilliant and very intriguing analysis. Came here through https://swprs.org/covid-19-virus-origin ... ypothesis/
They link to another worrying article: https://armswatch.com/project-g-2101-pe ... s-in-bats/
So “Eco Health Alliance” was working with both the WIV and the Pentagon.
Allison Wilson
JULY 17, 2020 AT 10:29 PM REPLY
To answer Philip Ward: Good question. Our best guess is that it took a while for virus to evolve to be highly transmissible from human to human — this most likely happened later, after the miner(s) were in hospital. Also, once the miners were in the hospital the thesis is clear they were treated as if they had infectious diseases — perhaps even SARS or other very deadly diseases. Thus we expect the hospital staff were taking every precaution. There are other possibilities — the others were infected but had no symptoms or that infections were not made public or that we just have not encountered the data yet.
Dob
JULY 18, 2020 AT 12:24 AM REPLY
Being a scientist but not a research scientist, it seems from the evidence so far that the miners were infected by an undeveloped strain of Covid-19. Even assuming that RaTG13 is natural and its postulated genome is correct and not a hypothesised extrapolation from the 370 nucleotide sample from BAT4991 (neither of which appears to have any surviving examples despite being listed as being used for research numerous times in 2017 and 2018 in the Chinese research database) there is ample scientific evidence in related research quoted in the above article that it would have taken between 20 -50 years for the RaTG13 genome to have mutated naturally to the C0vid-19 genome.
However, in 2013 previous coronoviruses had not had the human to human transmission of Covid-19 and you would not have expected doctors, nurses, ancilliary staff, let alone simple porters, stretcher bearers and good samaritans to have taken the infection precautions we now take automatically – and yet none appears to have been infected. This means at that time human to human transmission was not possible – because that part of the virus’s genome had not come to fruition.
From 2013 to 2019 various Gain of Function processes were actioned on Bat samples in the Wuhan lab to make the samples more infectious to humans in order, as the researchers are quick to explain, to enable vaccines or preventative infectious measures to become readily available. In seven years what sort of experiments could have been done to arrive at Covid-19 – look at what has been done just in the last 8 months to counter the infection. See Timothy Stout’s comments for the range of options.
What I would like to know is how the infections in Wuhan, mostly type B virus with some A and the infections in Guangdong, all type A tie up and the timeframes involved. Did researchers from Wuhan travel to Guangdong and infect locals and did the virus then mutate from A to B as A was less infectious in the Chinese hosts (possibly from previous coronavirus infections) and therefore the virus had to change. Much to be discussed.
Madeleine Love
JULY 18, 2020 AT 1:05 AM REPLY
Thank you so much for the effort to find and translate a very interesting thesis, and developing a new hypothesis.
I had questions on the time related epidemiology, and also on the question raised by Philip above. I have questions about the ability to extract full sequences some months after the infection event, based on government reporting from my home state.
The article read as though all miners passaged RaTG13 for the same outcome. One would think there’d be an infection precedent. On the hypothesis that the miners suffered from Sars-Cov-2, I would think that one would’ve passaged it first.
[The typcial reported course of the virus… There is average of 6 days from infection to mild symptoms (2-26?). Another week to symptoms becoming “serious” enough to require hospitalisation.]
Patients were admitted 26/4 (1), 25/4 (2), 27/4 (3), 26/4 (4), 2/5 (5), 26/4 (6). Patients (1)-(4) began at the mine on 2/4 and worked 2 weeks. Patients (5) and (6) began on 22/4, supposedly after (1)-(4) had left. I would ask questions about residence and contact. It’s possible the one of the first miners passaged the most successful virus, and working an enclosed space passed it to the others within a few days. Certainly patient (6) proceeded more rapidly to hospital (patient (5) quite quickly), so it would suggest they already had the virus. I’d be asking questions about where they lived – local or fly in/fly out? Do they live in a town with others? Is there any local report of doctors/hospital staff becoming infected? I suppose China had been through sars and were highly precautionary – this didn’t apply in Wuhan where there were many infections in medical staff, but perhaps this was a consequence of an avalanche of cases and low ppe availability. It’s said that 80% of cases pass the virus on to no-one and that 20% are the super-spreaders. Perhaps the 6 weren’t the spreading types?
If this was the Sars-Cov-2 virus, one would be looking for the greater prevalence of low symptom sufferers, understanding that the miners were a relatively unwell population.
Regarding the ability to extract full sequences some months after the infection event. Here in Victoria Australia there is a general view that people are non-infectious 10-14 days after first symptoms. They explain subsequent positive tests as being related to viral particles, not whole live viruses. They may not be right, or it may relate specifically to the tests they are using, but they lead me to ask questions about the extraction of full viral material from the thymus so long after the infection events.
Jonathan Latham
JULY 18, 2020 AT 12:18 PM REPLY
I hope I have understood your question correctly but our contention is that, while normal coronavirus passes through a person quickly, these miners had the disease continuously for months. They did not manage to clear it and there was a kind of stalemete between the virus and the immune system that took a long time to resolve, either in death or recovery. We don’t rule out that they may have passed it to each other. Jonathan
Madeleine Love
JULY 18, 2020 AT 4:46 AM REPLY
The fact that the miners were sick with Covid-19-like symptoms doesn’t preclude lab development to enhance human affinity. What they presented with may have been the symptoms of BtCoV/4991 and RaTG13. Although these coronaviruses may not have strong human affinity it may have been sufficient in damaged lungs. This could explain the lack of a public epidemic/health care worker report of illness. Are there any reports of anyone having been infected with BtCoV/4991 and RaTG13?
Aksel Fridstrøm
JULY 18, 2020 AT 1:41 PM REPLY
Thank you for a very interesting article, and for providing a link to my interview with Birger Sørensen in Minerva.
I have recently written two more articles on the works of Sørensen, you might find them interesting.
This one is about the struggle to publish his article:
https://www.minervanett.no/angus-dalgle ... cle/362519
The second one contains an upload of his full thesis:
https://www.minervanett.no/angus-dalgle ... rus/362529
Allison Wilson
JULY 18, 2020 AT 2:34 PM REPLY
Lance thank you for the link to Arms Watch and the excellent article connecting EcoHealth Alliance (which has links to the Gates Foundation) to the US Military. I hope many people will read this article and the Arms Watch website http://armswatch.com/. EcoHealth gets the vast majority of its money from the government but it also has corporate funding and foundation funding which it would be useful to know more about.
Brin
JULY 18, 2020 AT 4:28 PM REPLY
@Jonathan and Allison: Prof Balloux notes that “4 out of 7 known hCoVs have furin cleavage site”, including “common cold” viruses OC43 and HKU1. Is this correct or is there a difference to SARS2 and MERS that makes them special or requires “special explanation”?
https://twitter.com/BallouxFrancois/sta ... 5746233344
Allison Wilson
JULY 18, 2020 AT 4:31 PM REPLY
There may exist other reports on the Mojiang Mine Outbreak, or reports of people other than the miners being infected. However, the only reports we are aware of are 1. The Master’s thesis by Xu,Li (2013) 2. The Huang, Canping (2016) PhD thesis(chapter 3). 3. The Science news item “A New Killer Virus in China?” (this report only mentions the three miners who died) and 4. the short scientific report by Wu et al. 2014. The originals and/or translations of these documents are all linked to in the text and/or cited in the references of our article. We would like to hear about any other documented reports on this outbreak that people can find.
Brin
JULY 18, 2020 AT 4:35 PM REPLY
alright this seems to answers it: no other CoV has ACE2/FCS combination:
https://twitter.com/flavinkins/status/1 ... 3676516352
https://twitter.com/flavinkins/status/1 ... 7473904640
So FCS not unique in general but in hCov group 2B.
Crocodilite
JULY 18, 2020 AT 6:29 PM REPLY
This excellent piece has at least three key ideas or revelations. (1) The mine saga and human samples, a bombshell! (2) Significant and rapid adaptation by the virus in miner lungs. (3) ALL, not just some, features of the virus (including the all-important furin site) were acquired solely in miner lungs. Some may deem (2) and/or (3) controversial.
But one of the greatest contributions this piece could make is to alert people to a line of possible forensic inquiry that has a clear origin and narrative that should be pursued. We now have a solid lead, starting with the mine, to the hospital, to WIV. A lot of people were involved in this chain of events. There should be witnesses, paper trails, and medical samples. That these leads need to be pursued is entirely independent of whether one believes Idea (2) or Idea (3). The corrupt forces will likely want to confuse the matter. They would point to the potential controversies of Idea (2) or Idea (3) to discredit the entire piece and therefore obstruct an inquiry that starts with just Idea (1). People need to be aware of that and call that out if/when it happens.
And for people who believe an entirely man-made narrative, please be mindful that Idea (1) and Idea (2) are still compatible with later human manipulation. You wouldn’t want to go out of your way to discredit this piece just because you don’t believe in Idea (3). Unlike the disgraceful Andersen piece, this piece is your friend, not your enemy. The mine origin story could be one of the very few, if not the only, credible forensic leads that could be the starting point of an inquiry. If an investigation that starts with the mine ultimately leads to uncovering crimes that more closely resemble your pet theory than Idea (3), I believe the authors of this piece would have no problem, neither should you. We should be objective and critical about scientific details, but we should be united in our call for an inquiry that follows the trail leading from the mine, to the hospital, to WIV, and hopefully, to the ultimate truth!
Paul Jackson
JULY 18, 2020 AT 6:34 PM REPLY
How about the presence of a snippet of HIV in SARS-CoV-2? Does that exist? Does it point to some manipulation in the WIV lab?
Alan
JULY 19, 2020 AT 8:32 PM REPLY
The story is convincing in detail, but fails to support the conclusions. The story says that Covid19 evolved naturally in miners lungs in a hospital, then they concluded that it originated from a lab. According to the detail, the lab merely stored the virus and did no significant originating work on it before it escaped. this does NOT make the lab the origin of the virus, merely a multi-year storage point during which time it seems to have been eradicated elsewhere.
M
JULY 20, 2020 AT 7:06 AM REPLY
SARS-CoV-2, viruses that are suspected to have caused a COVID-like illness in 2012 and which may be key to understanding not just the origin of the COVID-19 pandemic but the future behaviour of SARS-CoV-2. Well said as Dr Yan limeng in the Fox News interview warn us we need find the truth.
Jonathan Latham
JULY 20, 2020 AT 7:47 PM REPLY
PS Apologies that the “REPLY” function seems not to be working. Attempting to fix this.
Jonathan Latham
JULY 22, 2020 AT 3:41 PM REPLY
A kind reader (John) has made this available as a Talking Paper to download and listen to. Hear his nice English accent at:
https://soundcloud.com/talkingpapers/pr ... r-sarscov2
Also available on youtube:
https://youtu.be/l3gQdkWn9u0
(link updated)
Paul Zhao
JULY 23, 2020 AT 9:38 AM REPLY
Would it be possible to publish the paper on mainstream science journals? If not, it’s unlikely to lead to an unbiased investigation, and the article will be ignored, as Dr. Sorensen’s.
Jonathan Latham
JULY 23, 2020 AT 12:40 PM REPLY
The conundrum of peer review is that it can take a long time and may not be fair. One of the things we have noticed in submitting ‘challenging’ papers is that editors (and/or reviewers) often take even longer than normal. It can take six months to get a “No” and that No is sometimes based on a trivial critique. We therefore don’t cite Sorensen just because he says it. We have personal experience. This paper of ours, for example ended up never being published even though no one (including peer-reviewers, editors, etc) has ever advanced a serious and substantive critique (https://2k4vbx44lajeo2rag2seu29o-wpengi ... Wilson.pdf).
But the other aspect is that, even if it becomes part of the published literature, it can still be ignored.
Lifeguard Larry
JULY 23, 2020 AT 11:01 PM REPLY
https://www.scientificamerican.com/arti ... onavirus1/
Notice
1) China says it has less infections and deaths now than it had in April
2) Shi says that the Mojiang miners suffered from a fungal infection when it was known almost immediately that they suffered from coronavirus.
3) Daszek is not willing to suspend his dangerous experiments despite the huge risks now evident to everyone.
4) Scientific American did not revise the numbers or comment on any of this when it republished the article. Nor has the scientific community subsequently.
Bob Clyatt
JULY 24, 2020 AT 4:51 PM REPLY
Following these papers and discussion with great interest. Thank you for your courageous and intellectually honest efforts. One question that I did not see addressed: what would explain the highly stable nature of Covid-19 in humans? Does that suggest some sort of passaging process might have been undertaken on the original miner’s samples at WIV during the past few years, and that one of those more evolved/stable versions, rather than the raw miner samples, was what leaked?
Will Lar
JULY 24, 2020 AT 7:45 PM REPLY
If it’s from the lab samples of WIV as suggested, it’s naturally to think that the workers especially the ones in the Shi lab in the institute were the first to be exposed and possibly infected. Do you have any information or data regarding the number of infected cases or deaths within WIV? Any information regarding the possible pathway of virus traveling from the lab to the outside world?
U.S. government gave $3.7 million grant to Wuhan lab at cent
Re: U.S. government gave $3.7 million grant to Wuhan lab at
The most logical explanation is that it comes from a laboratory: The well-known Norwegian virologist Birger Sørensen and his colleagues have examined the corona virus. They believe it has certain properties which would not evolve naturally. These conclusions are politically controversial, but in this interview he shares the findings behind the headlines.
by Aksel Fridstrøm
Translated from Norwegian by Kathrine Jebsen Moore
July 2, 2020 - 19:10 SIST OPPDATERT Søndag 19. juli 2020 - 09:56
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT

“I understand that this is controversial, but the public has a legitimate need to know, and it is important that it is possible to freely discuss alternate hypotheses on how the virus originated” Birger Sørensen starts to explain when Minerva visits him in his office one morning in Oslo.
Despite the explosiveness of his statements and research, Sørensen remains calm and collected.
Sørensen has been a point of controversy ever since former MI6 director Richard Dearlove cited a yet to be published article by Sørensen and his colleagues in an interview with The Daily Telegraph. The article claims that the virus that causes Covid-19 most likely has not emerged naturally.
“It’s a shame that there has already been so much talk about this, because I have yet to publish the article where I put forward my analysis”, Sørensen says in the form of an exasperated sigh.
Together with his colleagues, Angus Dalgleish and Andres Susrud have authored an article that looks into the most plausible explanations regarding the origins of the novel coronavirus. The article builds upon an already published article in the Quarterly Review of Biophysics that describes newly discovered properties in the virus spike protein. The authors are still in dialogue with scientific journals regarding an upcoming publication of the article.
News outlets are thus confronted with a difficult question: Are the findings and arguments Sørensen and his colleagues put forward of a sufficiently high quality to be presented and discussed in the public sphere? Sørensen explains that they in their dialogue with scientific journals are encountering a certain reluctance to publishing the article – without, however, proper scientific objections. Minerva has read a draft of the article, and has after an overall assessment decided that the findings and arguments do deserve public debate, and that this discussion cannot depend entirely on the publication process of scientific journals.
In this interview with Minerva, Sørensen therefore puts forward his hypothesis on why it is highly unlikely that the coronavirus emerged naturally.
On May 18th, WHO decided to conduct an inquiry into the coronavirus epidemic in China. Sørensen believes that it is important that this inquiry looks into new and alternate explanations for how the virus originated, beyond the already well-known suggestion that the virus originated in the Wuhan Seafood Market.
“There are very few who still believe that the epidemic started there, so as of today we have no good answers on how the epidemic started. Then we must also dare to look at more controversial, alternative explanations for the origin,” Sørensen says.
Birger Sørensen and one of his co-authors, Angus Dalgleish, are already known as HIV researchers par excellence.
In 2008, Sørensen’s work came to international attention when he launched a new immunotherapy for HIV. Angus Dalgleish is the professor at St. George’s Medical School in London who became world famous in 1984 after having discovered a novel receptor that the HIV virus uses to enter human cells.
The purpose of the work Sørensen and his colleagues have done on the novel coronavirus, has been to produce a vaccine. And they have taken their experience in trialling HIV vaccines with them to analyse the coronavirus more thoroughly, in order to make a vaccine that can protect against Covid-19 without major side effects.
Exceptionally well adjusted
“The difference between our approach and other vaccine manufacturers is that we have a chemistry background, and we analyse the virus in detail as if we were making a drug,” Sørensen starts to explain.
“Biology is also chemistry, so by considering the virus from a chemistry perspective, we carry out more detailed analysis, zooming in on certain components.”
Sørensen takes us through the basic elements of their approach:
“The first thing you need to establish is which parts of the virus are changing, and which parts are stable. If you want to make a vaccine that lasts, you must stimulate the immune system to react against those parts of the virus that are constant, otherwise the effect will disappear and, in the worst-case scenario, lead to increased illness.
“Once we know this, we can try to make a vaccine. Where we differ is that we are trying to make a vaccine that uses elements that have as little in common with the body’s natural components as possible, so that the immune system is taught to recognise exactly what the vaccine should protect against”, Sørensen elaborates.
Sørensen believes this is an important insight which will prevent the immune system from being falsely stimulated in a way that could lead the vaccine to create too many dangerous side effects in the vaccinated person.
“When we have not succeeded in creating an HIV vaccine, despite the enormous efforts put into that endeavour for the past 30 years, it is because we haven’t understood this,” Sørensen continues.
He believes that there has not been enough interaction between the part of the pharmaceutical industry that makes HIV medicines and the part that runs the vaccine research. As a consequence, the knowledge you need to make a successful vaccine against HIV in the big pharmaceutical companies has not been adequately exploited by the big, international HIV preventing vaccine studies that have been carried out.”
Asked about what significance his approached has had when he has analyzed the coronavirus, Sørensen explains:
He pauses for a second.
“So well that it was suspicious,” he adds.
Perfected to infect humans
It is already known that the novel coronavirus, like the virus that caused the SARS epidemic in Southeast Asia in 2002-2003, could attach itself to the ACE-2 receptors in the lower respiratory tract.
“But what we have discovered is that there are properties in this new virus which enables it to use an additional receptor, and create a binding to human cells in the upper respiratory tract and the intestines which is strong enough to produce an infection,” Sørensen elaborates.
Sørensen says that it is the use of this additional receptor that most likely results in a different illness in Covid-19 patients than the one resulting from SARS.
“This is what enables the virus to transmit to a greater degree between humans, without the virus having attached itself to the ACE-2 receptors in the lower respiratory tract, where it causes deep pneumonia.
“That is also why so many of the Covid-19 patients have mild symptoms at the start of the illness, and are contagious before they develop severe symptoms,” he adds.
It might also explain why some people are ‘super spreaders’ without being ill themselves, Sørensen says.
In the already published article Sørensen and his colleagues Angus Dalgleish and Andres Susrud describe what they claim is curious about the spike protein of the coronavirus, which makes it especially well suited to infect humans. These findings are the foundation for the hypothesis Sørensen and his colleagues develop in the new article, where they claim that the virus is not natural in origin.
“There are several factors that point towards this,” says Sørensen. “Firstly, this part of the virus is very stable; it mutates very little. That points to this virus as a fully developed, almost perfected virus for infecting humans.
“Secondly, this indicates that the structure of the virus cannot have evolved naturally. When we compare the novel coronavirus with the one that caused SARS, we see that there are altogether six inserts in this virus that stand out compared to other known SARS viruses,” he goes on explaining.
Sørensen says that several of these changes in the virus are unique, and that they do not exist in other known SARS coronaviruses.
“Four of these six changes have the property that they are suited to infect humans. This kind of aggregation of a type of property can be done simply in a laboratory, and helps to substantiate such an origin,” Sørensen points out.
An artificially created virus
Asked about whether this implies that the virus is not natural, Sørensen goes on to explain the laboratory process that leads to the creation of new viruses.
“In a sense it is natural. But the natural processes have most likely been accelerated in a laboratory,” he explains. “It’s also possible for a virus to attain these properties in nature, but it’s not likely. If the mutations had happened in nature, we would have most likely seen that the virus had attracted other properties through mutations, not just properties that help the virus to attach itself to human cells.”
Sørensen vividly explains this argument:
“Imagine that you have cultivated a billion coronaviruses you have gathered from nature, then you take this mass of viruses and inject them into a human cell culture from for example the upper respiratory tract. As a result, a few of these viruses will change in order to better attach themselves to this type of cell in the nose and throat region and therefore to infect humans more easily. You end up with a virus with a spike protein which is perfect for attaching to and penetrating human cells.” Sørensen explains.
Asked about the particular mutations in the virus that lead to this conclusion, Sørensens says:
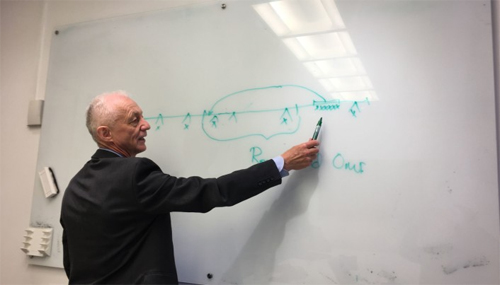
Sørensen explains at the black board. Photo: Aksel Fridstrøm
On a board in the meeting room where Sørensen is hosting our meeting, he illustrates what he is trying to explain, and how a component of the virus which previously was situated on another part of the shell of the virus, now has become a part of the spike protein of the virus.
More than ninety percent confident
Sørensen is therefore quite confident that the virus has originated in a laboratory.
“I think it’s more than 90 percent certain. It’s at least a far more probable explanation than it having developed this way in nature”, Sørensen responds.
Sørensen also highlights other data than those related to the virus’ properties:
“The properties that we now see in the virus, we have yet to discover anywhere in nature. We know that these properties make the virus very infectious, so if it came from nature, there should also be many animals infected with this, but we have still not been able to trace the virus in nature.
“The only place we are aware of where an equivalent virus to that which causes Covid-19 exists, is in a laboratory. So the simplest and most logical explanation is that it comes from a laboratory. Those who claim otherwise, have the burden of proof,” Sørensen says.
Critical voices suppressed
There are indeed earlier known experiments where changes to the corona virus have been engineered. An interesting example of this kind of research is a collaborative effort between Wuhan Institute of Virology and University of North Carolina at Chapel Hill. In a 2015 article, Menachery et al. describe experiments with laboratory created corona viruses -– so called gain-of-function-studies. The purpose of this research is partly to be better prepared for new pathogenic variants of the virus. But the researchers also write: "the potential to prepare for and mitigate future outbreaks must be weighed against the risk of creating more dangerous pathogens". This risk must also be evaluated in light of previous known accidents where corona viruses have escaped from laboratories in China.
But several researchers have already pointed out that artificially created viruses would be easy to identify. We therefore ask Sørensen why this has not been identified earlier.
Sørensen believes there are several reasons for this.
“The first is that this is a very uncomfortable finding, and the production of new scientific articles that can be used to prove such findings has all but ground to a halt. Chinese scientists no longer publish articles that can be used to support such a hypothesis”, he says.
“And newer articles that are published about the virus must be thoroughly investigated, especially in relation to the basic material that is being used,” Sørensen expands, and points to a new x-ray article published in Nature by Shang et al., which Sørensen also earlier has criticised for being misleading.
“To do my analysis, I have therefore had to go back to the source material, and look at those articles that were published before the Covid-19 outbreak, where we have chosen to assume that the data that have been used is okay and reflects the actual conditions,” Sørensen says.
Asked about why there has not been more debate on this topic Sørensen has several explanations.
“This quickly becomes a discussion on politics, rather than science, Sørensen responds.
“Nobody wants to put forward the inconvenient truth, many scientists are also concerned about their own funding and position if they were to put forward such a controversial hypothesis. It is nevertheless a fact that many people on the web have engaged in such a debate. But so far, those who participate in such forums are characterized as conspiratorial. It is also the case that a debate about this type of viral research and the technologies used may damage reputation and lead to new restrictions on how to conduct molecular genetic research. With this in mind, it is not difficult to see that it must be difficult to get accepted papers in peer reviewed journals that focus on such research", Sørensen elaborates.
Hopes his arguments will be discussed properly
Sørensens himself is Chairman of the board at Immunor, a company which is working to develop their own vaccine candidate for Covid-19. Minerva has challenged him to address allegations that this hypothesis is launched publicly to attract funding for his own research.
“Of course, it’s in my interest that my research becomes known, but I am being completely open and have declared all my interests.
“At the same time, I argue that it must be possible for those of us who work for smaller biotechnology companies to present our findings and get them discussed properly. If anyone wishes to contest my findings, they are of course welcome to do so, but I hope they will engage thoroughly with the arguments rather than derail them by discussing my motives,” Sørensen responds.
*****************************
The fight for a controversial article: Birger Sørensen and Angus Dalgleish failed to get an article about the origins of the coronavirus published in a scientific journal. The authors suspect foul play and political considerations. Not everything gets published, is the answer from the journals. Minerva has obtained a draft of the paper, to let readers and researchers decide.
by Aksel Fridstrøm
July 13 2020 - 16:45 SIST OPPDATERT Mandag 13. juli 2020 - 20:10
In an interview with Minerva last week, the well-known Norwegian vaccine researcher and physical chemist Birger Sørensen argued that the novel coronavirus is not natural in origin.
Together with his colleagues Angus Dalgleish and Andres Susrud -– the latter in a data analytical role as statistician and data miner –- Sørensen has written a series of journal articles that put forward arguments for why the most likely explanation for the origin of the coronavirus is a laboratory.
If such findings were confirmed, there could be political ramifications. Naturally, therefore, Sørensen, Dalgleish and their unpublished paper have been mired in controversy ever since Sir Richard Dearlove, former head of the MI6, endorsed their conclusions. The authors themselves suspect that the controversial conclusions and the heated debate may have made journals reluctant to evaluate their paper objectively. However, tons of scientific articles are rejected for any number of reasons.
By tracing the writing of articles, the contact between the authors and the journals, and reviewing the findings, this article aims to shed light on a troubling question for the scientific community during the Corona crisis: One the one hand, there is an overabundance of papers and findings of highly variable quality -– some of which fuel conspiracy theories. On the other hand, the question of the origin of the Corona virus has become a fraught political question, with the Chinese government clamping down on independent research, and president Donald Trump claiming that the virus originated in a Chinese lab without producing any evidence to back up the assertion.
With the consent of Mr. Sørensen and his co-authors (henceforth: Sørensen and Dalgleish), Minerva has obtained a full print of the article, to be read freely by our readers – and by scientists, who may then discuss and dissect the paper.
Searching for a vaccine
It started out with something less controversial: Originally, the authors were engaged in analysis of the virus with the aim to create a vaccine. The discoveries that the authors claim to be relevant for determining the origin of the virus came as a by-product of this research.
In fact, Sørensen and Dalgleish have managed to get a paper on the corona virus peer reviewed and published, in Quarterly Reviews of Biophysics Discovery. This article, which is more closely linked to their vaccine development, deals with observations of the virus and the receptors that the virus can attach to in humans. Sørensen and Dalgleish do indeed believe that these properties indicate a lab origin for the virus. However, the article itself avoids any mention of this implication of the discovered properties.
Originally, the findings which are now published in Quarterly Reviews of Biophysics Discovery were part of a more condensed article, which included the lab origin hypothesis. To make this argument, the three have now instead written a second article, “The Evidence which Suggests that This Is No Naturally Evolved Virus”, that puts forward their arguments on why they believe the virus is likely to be a laboratory construct, by combining insights from the first paper with what is known from lab work on corona viruses.
However, this second article is yet to be accepted by a scientific journal -– having been rejected at different times and formats by three leading journals. Sørensen states that he is presently in dialogue with other journals regarding publication. A third paper on a related topic also taken from the original argument, is yet to be submitted.
Endorsed and criticized
Before we move on, let’s take a look on some of the reactions from within the scientific community. Sørensen has received both severe criticism and partial support.
Professor Kristian Andersen at the department of immunology and microbiology at Scripps Research, a medical research facility in California, was lead author on the article “The proximal origin of SARS-CoV-2” where he states that his “analyses clearly show that SARS-CoV-2 is not a laboratory construct or a purposefully manipulated virus”.
Andersen last week told Sky news that Sørensen’s and Dalgleish’s work was “complete nonsense, unintelligible, and not even remotely scientific – leading the authors to make unfounded and unsupported conclusions about the origin of SARS-CoV-2”. However, Andersen has not had an opportunity to read the second, unpublished article.
Minerva has, however, shared that second article with Birgitta Åsjö, a professor emerita in virology at the university of Bergen. She is puzzled by Andersen’s comments, and rejects the notion that Sørensen’s and Dalgleish’ work is unscientific nonsense. “Andersen is harsh, I don’t know why he’s so harsh. I don’t want to dismiss it completely”, she told Minerva in an interview.
Åsjö does not consider the article to contain conclusive evidence that the novel coronavirus has a lab origin, but adds that she considers the article to contain “interesting findings”. She is herself critical of some of the arguments presented in Andersen’s own article on the origin of the virus.
In particular, Åsjö is interested in Sørensen’s and Dalgleish’ findings concerning properties of a swine corona virus detected in connection with an outbreak among piglets in Guandong in 2016–2017, and its possible significance for the present SARS-CoV-2 virus. So far, the controversy seems to resemble other controversies between academics who rarely hesitate to describe rivalling theories in the harshest possible terms. Indeed, Sørensen and Dalgleish originally intended to publish their arguments as a stark critique of the article by Andersen et al in Nature and what they describe “as puzzling errors in their use of evidence”.
Self-confident scientists and strict journals
With this aim, Angus Dalgleish wrote a letter to the editor in Nature on April 2, requesting that journal to publish an earlier version of these arguments.
This article was rejected by Nature five days later, on April 7. Announcing the rejection, João Monteiro, chief editor in Nature Medicine, wrote to Dalgleish:
Monteiro ended the email by encouraging Dalgleish to post his comments in one of “the accepted preprint repositories so that it remains visible and adds to the discussion about the origin of the virus.”
A clearly angered Dalgleish then wrote a response stating: “Thank you for your extraordinarily unhelpful replies. We can only conclude that the Nature editorial team does not understand that there is no scientific issue in the world at present more important than establishing with scientific precision the aetiology of the Covid-19 virus.”
After the first rejection by Nature, the authors approached another premier journal, Journal of Virology. However, by April 20, the first version of the paper had been rejected there as well. A few days later, this version of the paper was put to death by a rejection from bioRxiv, a non-peer-reviewed preprint repository. The stated reason for rejection was that the format of the paper did not conform to a normal, full research paper, with sections such as "Methods" and "Results".
The first iteration of rejections thus seem to fit into a typical pattern: Scientists with overconfidence not only in the quality of their own research, but also its relevance and significance, encountering journals with strict guidelines for format, each with its own mission and focus, and not very patient with professors that flaunt formal requirements.
Still, it seems that the actual arguments put forward might not have been properly evaluated, or could not be properly evaluated in this setting. And the findings, if correct, would seem to merit some sort of scientific attention. How to proceed?
A publication and new rejections
After the initial round of rejections, the authors made several revisions to their original article, with the arguments sectioned into separate articles. The first article –- an analysis of the novel coronavirus, for the purpose of vaccine design, without making the argument that the virus is engineered –- was published June 2 in the Quarterly Reviews of Biophysics Discovery which is one of the top peer-reviewed science journals in the world.
Having achieved this publication, and presumably regained confidence that the scientific quality of the work was all right, Sørensen and Dalgleish again reached out to several of the world’s most prestigious scientific journals. Now they wanted to publish the second article, which builds on the first, already accepted article, and presents their arguments for why the coronavirus is of a non-natural origin.
However this article was again rejected by Nature on June 24 – without being sent out to peer review. The rejection, written by Senior Editor Clare Thomas, states:
On July 1, Sørensen and his colleagues therefore challenged Science, another scientific journal to publish his article. Arguing for the publication of the article Sørensen wrote in an e-mail to editor Professor Holden Thorp:
“Now that Dr. Tedros Adhanom Ghebreyesus has indicated that WHO will pursue a long-overdue inquiry into the aetiology of the SARS-CoV-2 virus, which we welcome, we hope that you will support the very necessary debate that is now breaking in a second wave, following the publication of our vaccine. We are aware of significant responsible mainstream media interventions that are imminent. We are glad that this important question will now be addressed where it should be, in mainstream media and science journals, and not left to internet speculations, some of which have been both uninformed and therefore unhelpful, in our view.”
However, Sørensen was rebuffed also by Science the very next day. In an email to Sørensen, Professor Throp wrote that the article was unfit for publishing in Science, due to the fact that it criticizes work published in another journal.
On the rejections from the scientific journals co-author Angus Dalgleish told Sky News: “I thought the whole point of a scientific journal was that you put forward some speculation and you opened it up to debate”, said Professor Dalgleish.
Agree or disagree with Dalgleish’s description of what a scientific journal does, the new round of rejections complicate the picture from the first round. A major part of the argument was accepted by a respectable journal -– the one that didn’t spell out the implications for the origin of the virus. The article that did spell it out, was now rejected twice, without peer review, and the second rejection on purely formal grounds that the authors vehemently contest, arguing that the paper in its current form is not first and foremost a critique of the Andersen et al. paper.
Minerva has asked both Nature and Science to elaborate on the reasons for why the articles by Sørensen and his co-authors have been rejected. Both Science and Nature have declined to comment on the specific rejection of the article as they view this information as confidential. However, Executive Director of the Science Press Package Meagan Phelan, replied that “Science receives upwards of 11,000 manuscripts per year, and the acceptance rate is 6%, so the vast majority of papers are rejected for one reason or another. Science’s acceptance rate for COVID-19-related submissions is even lower, at 4%. The journal continues to receive an exceptionally high volume of COVID-19-related submissions each week.”
At press time, the second article is still in search of a scientific journal willing to publish – or, at least, to subject it to peer review.
Correction: The name Quarterly Reviews of Biophysics has been changed to Quarterly Reviews of Biophysics Discovery
by Aksel Fridstrøm
Translated from Norwegian by Kathrine Jebsen Moore
July 2, 2020 - 19:10 SIST OPPDATERT Søndag 19. juli 2020 - 09:56
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
YOU ARE REQUIRED TO READ THE COPYRIGHT NOTICE AT THIS LINK BEFORE YOU READ THE FOLLOWING WORK, THAT IS AVAILABLE SOLELY FOR PRIVATE STUDY, SCHOLARSHIP OR RESEARCH PURSUANT TO 17 U.S.C. SECTION 107 AND 108. IN THE EVENT THAT THE LIBRARY DETERMINES THAT UNLAWFUL COPYING OF THIS WORK HAS OCCURRED, THE LIBRARY HAS THE RIGHT TO BLOCK THE I.P. ADDRESS AT WHICH THE UNLAWFUL COPYING APPEARED TO HAVE OCCURRED. THANK YOU FOR RESPECTING THE RIGHTS OF COPYRIGHT OWNERS.

“I understand that this is controversial, but the public has a legitimate need to know, and it is important that it is possible to freely discuss alternate hypotheses on how the virus originated” Birger Sørensen starts to explain when Minerva visits him in his office one morning in Oslo.
Despite the explosiveness of his statements and research, Sørensen remains calm and collected.
Sørensen has been a point of controversy ever since former MI6 director Richard Dearlove cited a yet to be published article by Sørensen and his colleagues in an interview with The Daily Telegraph. The article claims that the virus that causes Covid-19 most likely has not emerged naturally.
“It’s a shame that there has already been so much talk about this, because I have yet to publish the article where I put forward my analysis”, Sørensen says in the form of an exasperated sigh.
Together with his colleagues, Angus Dalgleish and Andres Susrud have authored an article that looks into the most plausible explanations regarding the origins of the novel coronavirus. The article builds upon an already published article in the Quarterly Review of Biophysics that describes newly discovered properties in the virus spike protein. The authors are still in dialogue with scientific journals regarding an upcoming publication of the article.
News outlets are thus confronted with a difficult question: Are the findings and arguments Sørensen and his colleagues put forward of a sufficiently high quality to be presented and discussed in the public sphere? Sørensen explains that they in their dialogue with scientific journals are encountering a certain reluctance to publishing the article – without, however, proper scientific objections. Minerva has read a draft of the article, and has after an overall assessment decided that the findings and arguments do deserve public debate, and that this discussion cannot depend entirely on the publication process of scientific journals.
In this interview with Minerva, Sørensen therefore puts forward his hypothesis on why it is highly unlikely that the coronavirus emerged naturally.
On May 18th, WHO decided to conduct an inquiry into the coronavirus epidemic in China. Sørensen believes that it is important that this inquiry looks into new and alternate explanations for how the virus originated, beyond the already well-known suggestion that the virus originated in the Wuhan Seafood Market.
“There are very few who still believe that the epidemic started there, so as of today we have no good answers on how the epidemic started. Then we must also dare to look at more controversial, alternative explanations for the origin,” Sørensen says.
Birger Sørensen and one of his co-authors, Angus Dalgleish, are already known as HIV researchers par excellence.
In 2008, Sørensen’s work came to international attention when he launched a new immunotherapy for HIV. Angus Dalgleish is the professor at St. George’s Medical School in London who became world famous in 1984 after having discovered a novel receptor that the HIV virus uses to enter human cells.
The purpose of the work Sørensen and his colleagues have done on the novel coronavirus, has been to produce a vaccine. And they have taken their experience in trialling HIV vaccines with them to analyse the coronavirus more thoroughly, in order to make a vaccine that can protect against Covid-19 without major side effects.
Exceptionally well adjusted
“The difference between our approach and other vaccine manufacturers is that we have a chemistry background, and we analyse the virus in detail as if we were making a drug,” Sørensen starts to explain.
“Biology is also chemistry, so by considering the virus from a chemistry perspective, we carry out more detailed analysis, zooming in on certain components.”
Sørensen takes us through the basic elements of their approach:
“The first thing you need to establish is which parts of the virus are changing, and which parts are stable. If you want to make a vaccine that lasts, you must stimulate the immune system to react against those parts of the virus that are constant, otherwise the effect will disappear and, in the worst-case scenario, lead to increased illness.
“Once we know this, we can try to make a vaccine. Where we differ is that we are trying to make a vaccine that uses elements that have as little in common with the body’s natural components as possible, so that the immune system is taught to recognise exactly what the vaccine should protect against”, Sørensen elaborates.
Sørensen believes this is an important insight which will prevent the immune system from being falsely stimulated in a way that could lead the vaccine to create too many dangerous side effects in the vaccinated person.
“When we have not succeeded in creating an HIV vaccine, despite the enormous efforts put into that endeavour for the past 30 years, it is because we haven’t understood this,” Sørensen continues.
He believes that there has not been enough interaction between the part of the pharmaceutical industry that makes HIV medicines and the part that runs the vaccine research. As a consequence, the knowledge you need to make a successful vaccine against HIV in the big pharmaceutical companies has not been adequately exploited by the big, international HIV preventing vaccine studies that have been carried out.”
Asked about what significance his approached has had when he has analyzed the coronavirus, Sørensen explains:
“We have examined which components of the virus are especially well suited to attach themselves to cells in humans. And we have done this by comparing the properties of the virus with human genetics. What we found was that this virus was exceptionally well adjusted to infect humans.”
He pauses for a second.
“So well that it was suspicious,” he adds.
Perfected to infect humans
It is already known that the novel coronavirus, like the virus that caused the SARS epidemic in Southeast Asia in 2002-2003, could attach itself to the ACE-2 receptors in the lower respiratory tract.
“But what we have discovered is that there are properties in this new virus which enables it to use an additional receptor, and create a binding to human cells in the upper respiratory tract and the intestines which is strong enough to produce an infection,” Sørensen elaborates.
Sørensen says that it is the use of this additional receptor that most likely results in a different illness in Covid-19 patients than the one resulting from SARS.
“This is what enables the virus to transmit to a greater degree between humans, without the virus having attached itself to the ACE-2 receptors in the lower respiratory tract, where it causes deep pneumonia.
“That is also why so many of the Covid-19 patients have mild symptoms at the start of the illness, and are contagious before they develop severe symptoms,” he adds.
It might also explain why some people are ‘super spreaders’ without being ill themselves, Sørensen says.
In the already published article Sørensen and his colleagues Angus Dalgleish and Andres Susrud describe what they claim is curious about the spike protein of the coronavirus, which makes it especially well suited to infect humans. These findings are the foundation for the hypothesis Sørensen and his colleagues develop in the new article, where they claim that the virus is not natural in origin.
“There are several factors that point towards this,” says Sørensen. “Firstly, this part of the virus is very stable; it mutates very little. That points to this virus as a fully developed, almost perfected virus for infecting humans.
“Secondly, this indicates that the structure of the virus cannot have evolved naturally. When we compare the novel coronavirus with the one that caused SARS, we see that there are altogether six inserts in this virus that stand out compared to other known SARS viruses,” he goes on explaining.
Sørensen says that several of these changes in the virus are unique, and that they do not exist in other known SARS coronaviruses.
“Four of these six changes have the property that they are suited to infect humans. This kind of aggregation of a type of property can be done simply in a laboratory, and helps to substantiate such an origin,” Sørensen points out.
FACT BOX – Spike Protein
A spike protein is a part of the virus attached to the surface of the virus. The spike protein is used by the virus when it enters cells, enabling it to stick in humans. The properties of the spike determines which receptors a virus can utilise and thus which cells the virus can enter to create illness.
An artificially created virus
Asked about whether this implies that the virus is not natural, Sørensen goes on to explain the laboratory process that leads to the creation of new viruses.
“In a sense it is natural. But the natural processes have most likely been accelerated in a laboratory,” he explains. “It’s also possible for a virus to attain these properties in nature, but it’s not likely. If the mutations had happened in nature, we would have most likely seen that the virus had attracted other properties through mutations, not just properties that help the virus to attach itself to human cells.”
Sørensen vividly explains this argument:
“Imagine that you have cultivated a billion coronaviruses you have gathered from nature, then you take this mass of viruses and inject them into a human cell culture from for example the upper respiratory tract. As a result, a few of these viruses will change in order to better attach themselves to this type of cell in the nose and throat region and therefore to infect humans more easily. You end up with a virus with a spike protein which is perfect for attaching to and penetrating human cells.” Sørensen explains.
Asked about the particular mutations in the virus that lead to this conclusion, Sørensens says:
“What we see is that an area that you could observe in the first SARS coronavirus has been moved, so that the parts of the virus that are particularly well suited to attach to humans, have become part of the spike protein that the virus uses to penetrate human cells. And it is this moving of the area of the virus which makes the virus, together with the injected areas explained above, able to utilise an additional receptor to infect humans.”

Sørensen explains at the black board. Photo: Aksel Fridstrøm
On a board in the meeting room where Sørensen is hosting our meeting, he illustrates what he is trying to explain, and how a component of the virus which previously was situated on another part of the shell of the virus, now has become a part of the spike protein of the virus.
More than ninety percent confident
Sørensen is therefore quite confident that the virus has originated in a laboratory.
“I think it’s more than 90 percent certain. It’s at least a far more probable explanation than it having developed this way in nature”, Sørensen responds.
Sørensen also highlights other data than those related to the virus’ properties:
“The properties that we now see in the virus, we have yet to discover anywhere in nature. We know that these properties make the virus very infectious, so if it came from nature, there should also be many animals infected with this, but we have still not been able to trace the virus in nature.
“The only place we are aware of where an equivalent virus to that which causes Covid-19 exists, is in a laboratory. So the simplest and most logical explanation is that it comes from a laboratory. Those who claim otherwise, have the burden of proof,” Sørensen says.
Critical voices suppressed
There are indeed earlier known experiments where changes to the corona virus have been engineered. An interesting example of this kind of research is a collaborative effort between Wuhan Institute of Virology and University of North Carolina at Chapel Hill. In a 2015 article, Menachery et al. describe experiments with laboratory created corona viruses -– so called gain-of-function-studies. The purpose of this research is partly to be better prepared for new pathogenic variants of the virus. But the researchers also write: "the potential to prepare for and mitigate future outbreaks must be weighed against the risk of creating more dangerous pathogens". This risk must also be evaluated in light of previous known accidents where corona viruses have escaped from laboratories in China.
Fact box: Gain-of-function studies
According to US Department health & human services, gain-of-function studies refer to research which aims to increase the ability of a pathogen to cause disease. The method is controversial, because it entails risks, such as viruses escaping from labs. Between 2014 and 2018, this kind of research was prohibited in the United States, but in December 2017, American authorities announced that the ban would be lifted.
But several researchers have already pointed out that artificially created viruses would be easy to identify. We therefore ask Sørensen why this has not been identified earlier.
Sørensen believes there are several reasons for this.
“The first is that this is a very uncomfortable finding, and the production of new scientific articles that can be used to prove such findings has all but ground to a halt. Chinese scientists no longer publish articles that can be used to support such a hypothesis”, he says.
“And newer articles that are published about the virus must be thoroughly investigated, especially in relation to the basic material that is being used,” Sørensen expands, and points to a new x-ray article published in Nature by Shang et al., which Sørensen also earlier has criticised for being misleading.
“To do my analysis, I have therefore had to go back to the source material, and look at those articles that were published before the Covid-19 outbreak, where we have chosen to assume that the data that have been used is okay and reflects the actual conditions,” Sørensen says.
Asked about why there has not been more debate on this topic Sørensen has several explanations.
“This quickly becomes a discussion on politics, rather than science, Sørensen responds.
“Nobody wants to put forward the inconvenient truth, many scientists are also concerned about their own funding and position if they were to put forward such a controversial hypothesis. It is nevertheless a fact that many people on the web have engaged in such a debate. But so far, those who participate in such forums are characterized as conspiratorial. It is also the case that a debate about this type of viral research and the technologies used may damage reputation and lead to new restrictions on how to conduct molecular genetic research. With this in mind, it is not difficult to see that it must be difficult to get accepted papers in peer reviewed journals that focus on such research", Sørensen elaborates.
The fight for a controversial article
Birger Sørensen and Angus Dalgleish failed to get an article about the origins of the coronavirus published in a scientific journal. The authors suspect foul play and political considerations. Not everything gets published, is the answer from the journals. Minerva has obtained a draft of the paper, to let readers and researchers decide.
Hopes his arguments will be discussed properly
Sørensens himself is Chairman of the board at Immunor, a company which is working to develop their own vaccine candidate for Covid-19. Minerva has challenged him to address allegations that this hypothesis is launched publicly to attract funding for his own research.
“Of course, it’s in my interest that my research becomes known, but I am being completely open and have declared all my interests.
“At the same time, I argue that it must be possible for those of us who work for smaller biotechnology companies to present our findings and get them discussed properly. If anyone wishes to contest my findings, they are of course welcome to do so, but I hope they will engage thoroughly with the arguments rather than derail them by discussing my motives,” Sørensen responds.
*****************************
The fight for a controversial article: Birger Sørensen and Angus Dalgleish failed to get an article about the origins of the coronavirus published in a scientific journal. The authors suspect foul play and political considerations. Not everything gets published, is the answer from the journals. Minerva has obtained a draft of the paper, to let readers and researchers decide.
by Aksel Fridstrøm
July 13 2020 - 16:45 SIST OPPDATERT Mandag 13. juli 2020 - 20:10
In an interview with Minerva last week, the well-known Norwegian vaccine researcher and physical chemist Birger Sørensen argued that the novel coronavirus is not natural in origin.
Together with his colleagues Angus Dalgleish and Andres Susrud -– the latter in a data analytical role as statistician and data miner –- Sørensen has written a series of journal articles that put forward arguments for why the most likely explanation for the origin of the coronavirus is a laboratory.
If such findings were confirmed, there could be political ramifications. Naturally, therefore, Sørensen, Dalgleish and their unpublished paper have been mired in controversy ever since Sir Richard Dearlove, former head of the MI6, endorsed their conclusions. The authors themselves suspect that the controversial conclusions and the heated debate may have made journals reluctant to evaluate their paper objectively. However, tons of scientific articles are rejected for any number of reasons.
By tracing the writing of articles, the contact between the authors and the journals, and reviewing the findings, this article aims to shed light on a troubling question for the scientific community during the Corona crisis: One the one hand, there is an overabundance of papers and findings of highly variable quality -– some of which fuel conspiracy theories. On the other hand, the question of the origin of the Corona virus has become a fraught political question, with the Chinese government clamping down on independent research, and president Donald Trump claiming that the virus originated in a Chinese lab without producing any evidence to back up the assertion.
With the consent of Mr. Sørensen and his co-authors (henceforth: Sørensen and Dalgleish), Minerva has obtained a full print of the article, to be read freely by our readers – and by scientists, who may then discuss and dissect the paper.
Searching for a vaccine
It started out with something less controversial: Originally, the authors were engaged in analysis of the virus with the aim to create a vaccine. The discoveries that the authors claim to be relevant for determining the origin of the virus came as a by-product of this research.
In fact, Sørensen and Dalgleish have managed to get a paper on the corona virus peer reviewed and published, in Quarterly Reviews of Biophysics Discovery. This article, which is more closely linked to their vaccine development, deals with observations of the virus and the receptors that the virus can attach to in humans. Sørensen and Dalgleish do indeed believe that these properties indicate a lab origin for the virus. However, the article itself avoids any mention of this implication of the discovered properties.
Originally, the findings which are now published in Quarterly Reviews of Biophysics Discovery were part of a more condensed article, which included the lab origin hypothesis. To make this argument, the three have now instead written a second article, “The Evidence which Suggests that This Is No Naturally Evolved Virus”, that puts forward their arguments on why they believe the virus is likely to be a laboratory construct, by combining insights from the first paper with what is known from lab work on corona viruses.
However, this second article is yet to be accepted by a scientific journal -– having been rejected at different times and formats by three leading journals. Sørensen states that he is presently in dialogue with other journals regarding publication. A third paper on a related topic also taken from the original argument, is yet to be submitted.
Endorsed and criticized
Before we move on, let’s take a look on some of the reactions from within the scientific community. Sørensen has received both severe criticism and partial support.
Professor Kristian Andersen at the department of immunology and microbiology at Scripps Research, a medical research facility in California, was lead author on the article “The proximal origin of SARS-CoV-2” where he states that his “analyses clearly show that SARS-CoV-2 is not a laboratory construct or a purposefully manipulated virus”.
Andersen last week told Sky news that Sørensen’s and Dalgleish’s work was “complete nonsense, unintelligible, and not even remotely scientific – leading the authors to make unfounded and unsupported conclusions about the origin of SARS-CoV-2”. However, Andersen has not had an opportunity to read the second, unpublished article.
The principal reason for media dismissals of the lab origin possibility is a review paper in Nature Medicine (Andersen et al., 2020). Although by Jun 29, 2020 this review had almost 700 citations it also has major scientific shortcomings. These flaws are worth understanding in their own right but they are also useful background for understanding the implications of the Master’s thesis.
Andersen et al., a critique
The question of the origin of the COVID-19 pandemic is, in outline, simple. There are two incontrovertible facts. One, the disease is caused by a human viral pathogen, SARS-CoV-2, first identified in Wuhan in December 2019 and whose RNA genome sequence is known. Second, all of its nearest known relatives come from bats. Beyond any reasonable doubt SARS-CoV-2 evolved from an ancestral bat virus. The task the Nature Medicine authors set for themselves was to establish the relative merits of each of the various possible routes (lab vs natural) by which a bat coronavirus might have jumped to humans and in the same process have acquired an unusual furin site and a spike protein having very high affinity for the human ACE2 receptor.
When Andersen et al. outline a natural zoonotic pathway they speculate extensively about how the leap might have occurred. In particular they elaborate on a proposed residence in intermediate animals, likely pangolins. For example, “The presence in pangolins of an RBD [Receptor Binding Domain] very similar to that of SARS-CoV-2 means that we can infer that this was probably in the virus that jumped to humans. This leaves the insertion of [a] polybasic cleavage site to occur during human-to-human transmission.” This viral evolution occurred in “Malayan pangolins illegally imported into Guangdong province”. Even with these speculations there are major gaps in this theory. For example, why is the virus so well adapted to humans? Why Wuhan, which is 1,000 Km from Guangdong? (See map).
The authors provide no such speculations in favour of the lab accident thesis, only speculation against it:
“Finally, the generation of the predicted O-linked glycans is also unlikely to have occurred due to cell-culture passage, as such features suggest the involvement of an immune system.” (italics added).
[Passaging is the deliberate placing of live viruses into cells or organisms to which they are NOT adapted for the purpose of making them adapted, i.e. speeding up their evolution.]
It is also noteworthy that the Andersen authors set a higher hurdle for the lab thesis than the zoonotic thesis. In their account, the lab thesis is required to explain all of the evolution of SARS-CoV-2 from its presumed bat viral ancestor, whereas under their telling of the zoonotic thesis the key step of the addition of the furin site is allowed to happen in humans and is thus effectively unexplained.
A further imbalance is that key information needed to judge the merits of a lab origin theory is missing from their account. As we detailed in our previous article, in their search for SARS-like viruses with zoonotic spillover potential, researchers at the WIV have passaged live bat viruses in monkey and human cells (Wang et al., 2019). They have also performed many recombinant experiments with diverse bat coronaviruses (Ge et al., 2013; Menachery et al., 2015; Hu et al., 2017). Such experiments have generated international concern over the possible creation of potential pandemic viruses (Lipsitch, 2018). As we showed too, the Shi lab had also won a grant to extend that work to whole live animals. They planned “virus infection experiments across a range of cell cultures from different species and humanized mice” with recombinant bat coronaviruses. Yet Andersen et al did not discuss this research at all, except to say:
“Basic research involving passage of bat SARS-CoV-like coronaviruses in cell culture and/or animal models has been ongoing for many years in biosafety level 2 laboratories across the world”
This statement is fundamentally misleading about the kind of research performed at the Shi lab.
A further important oversight by the Andersen authors concerns the history of lab outbreaks of viral pathogens. They write: “there are documented instances of laboratory escapes of SARS-CoV”. This is a rather matter-of-fact allusion to the fact that since 2003 there have been six documented outbreaks of SARS from labs, not all in China, with some leading to fatalities (Furmanski, 2014).
Andersen et al might have also have noted that two major human pandemics are widely accepted to have been caused by lab outbreaks of viral pathogens, H1N1 in 1977 and Venezuelan Equine Encephalitis (summarised in Furmanski, 2014). Andersen could even have noted that literally hundreds of lab accidents with viruses have resulted in near-misses or very localised outbreaks (summarised by Lynn Klotz and Sam Husseini and also Weiss et al., 2015).
Also unmentioned were instances where a lab outbreak of an experimental or engineered virus has been plausibly theorised but remains uninvestigated. For example, the most coherent explanation for the H1N1 variant ‘swine flu’ pandemic of 2009/10 that resulted in a death toll estimated by some as high as 200,000 (Duggal et al., 2016; Simonsen et al. 2013), is that a vaccine was improperly inactivated by its maker (Gibbs et al., 2009). If so, H1N1 emerged from a lab not once but twice.
Given that human and livestock viral outbreaks have frequently come from laboratories and that many scientists have warned of probable lab escapes (Lipsitch and Galvani, 2014), and that the WIV [Wuhan Institute of Virology] itself has a questionable biosafety record, the Andersen paper is not an even-handed treatment of the possible origins of the COVID-19 virus.
Yet its text expresses some strong opinions: “Our analyses clearly show that SARS-CoV-2 is not a laboratory construct or a purposefully manipulated virus….It is improbable that SARS-CoV-2 emerged through laboratory manipulation of a related SARS-CoV-like coronavirus…..the genetic data irrefutably show that SARS-CoV-2 is not derived from any previously used backbone….the evidence shows that SARS-CoV2 is not a purposefully manipulated virus….we do not believe that any type of laboratory-based scenario is possible.” (Andersen et al., 2020).
It is hard not to conclude that what their paper mostly shows is that Drs. Andersen, Rambaut, Lipkin, Holmes and Garry much prefer the natural zoonotic transfer thesis. Their rhetoric is forthright but the evidence does not support that confidence.
Indeed, since the publication of Andersen et al., important new evidence has emerged that undermines their zoonotic origin theory. On May 26th the Chinese CDC ruled out the Huanan “wet” market in Wuhan as the source of the outbreak. Additionally, new research on pangolins, the favoured intermediate mammal host, suggests they are not a natural reservoir of coronaviruses (Lee et al., 2020; Chan and Zhan, 2020). Furthermore, SARS-CoV-2 was found not to replicate in bat kidney or lung cells (Rhinolophus sinicus), implying that SARS-CoV-2 is not a recently-adapted spill over Chu et al., 2020)...
The final point that we would like to make is that the principal zoonotic origin thesis is the one proposed by Andersen et al. Apart from being poorly supported this thesis is very complex. It requires two species jumps, at least two recombination events between quite distantly related coronaviruses and the physical transfer of a pangolin (having a coronavirus infection) from outside China (Andersen et al., 2020). Even then it provides no logical explanation of the adaptedness of SARS-CoV-2 across its whole genome or why the virus emerged in Wuhan.
-- A Proposed Origin for SARS-CoV-2 and the COVID-19 Pandemic, by Jonathan Latham, PhD and Allison Wilson, PhD
Minerva has, however, shared that second article with Birgitta Åsjö, a professor emerita in virology at the university of Bergen. She is puzzled by Andersen’s comments, and rejects the notion that Sørensen’s and Dalgleish’ work is unscientific nonsense. “Andersen is harsh, I don’t know why he’s so harsh. I don’t want to dismiss it completely”, she told Minerva in an interview.
Åsjö does not consider the article to contain conclusive evidence that the novel coronavirus has a lab origin, but adds that she considers the article to contain “interesting findings”. She is herself critical of some of the arguments presented in Andersen’s own article on the origin of the virus.
In particular, Åsjö is interested in Sørensen’s and Dalgleish’ findings concerning properties of a swine corona virus detected in connection with an outbreak among piglets in Guandong in 2016–2017, and its possible significance for the present SARS-CoV-2 virus. So far, the controversy seems to resemble other controversies between academics who rarely hesitate to describe rivalling theories in the harshest possible terms. Indeed, Sørensen and Dalgleish originally intended to publish their arguments as a stark critique of the article by Andersen et al in Nature and what they describe “as puzzling errors in their use of evidence”.
Self-confident scientists and strict journals
With this aim, Angus Dalgleish wrote a letter to the editor in Nature on April 2, requesting that journal to publish an earlier version of these arguments.
This article was rejected by Nature five days later, on April 7. Announcing the rejection, João Monteiro, chief editor in Nature Medicine, wrote to Dalgleish:
First rejection from Nature Medicine
“While we appreciate that the points you have raised extend the discussion about the origin of SARS-CoV2 further our opinion is that the content complements other viewpoints that have been considered and published elsewhere, and therefore would be as appropriate for publication in the specialized literature. Please note that this is not a criticism regarding the importance of the matter or the quality of your analyses, but rather an editorial assessment of priority for publication, in a time when there are many pressing issues of public health and clinical interest that take precedence for publication in Nature Medicine, and limited space in the journal.”
Monteiro ended the email by encouraging Dalgleish to post his comments in one of “the accepted preprint repositories so that it remains visible and adds to the discussion about the origin of the virus.”
A clearly angered Dalgleish then wrote a response stating: “Thank you for your extraordinarily unhelpful replies. We can only conclude that the Nature editorial team does not understand that there is no scientific issue in the world at present more important than establishing with scientific precision the aetiology of the Covid-19 virus.”
After the first rejection by Nature, the authors approached another premier journal, Journal of Virology. However, by April 20, the first version of the paper had been rejected there as well. A few days later, this version of the paper was put to death by a rejection from bioRxiv, a non-peer-reviewed preprint repository. The stated reason for rejection was that the format of the paper did not conform to a normal, full research paper, with sections such as "Methods" and "Results".
The first iteration of rejections thus seem to fit into a typical pattern: Scientists with overconfidence not only in the quality of their own research, but also its relevance and significance, encountering journals with strict guidelines for format, each with its own mission and focus, and not very patient with professors that flaunt formal requirements.
Still, it seems that the actual arguments put forward might not have been properly evaluated, or could not be properly evaluated in this setting. And the findings, if correct, would seem to merit some sort of scientific attention. How to proceed?
A publication and new rejections
After the initial round of rejections, the authors made several revisions to their original article, with the arguments sectioned into separate articles. The first article –- an analysis of the novel coronavirus, for the purpose of vaccine design, without making the argument that the virus is engineered –- was published June 2 in the Quarterly Reviews of Biophysics Discovery which is one of the top peer-reviewed science journals in the world.
Having achieved this publication, and presumably regained confidence that the scientific quality of the work was all right, Sørensen and Dalgleish again reached out to several of the world’s most prestigious scientific journals. Now they wanted to publish the second article, which builds on the first, already accepted article, and presents their arguments for why the coronavirus is of a non-natural origin.
However this article was again rejected by Nature on June 24 – without being sent out to peer review. The rejection, written by Senior Editor Clare Thomas, states:
Second rejection from Nature Medicine
It is our policy to decline a substantial proportion of manuscripts without sending them to referees so that they may be sent elsewhere without further delay. In making this decision, we are not questioning the technical quality or validity of your findings, or their value to others working in this area, only assessing the suitability of the study based on the editorial criteria of the journal. In this case, we do not believe that the work represents a development of sufficient scientific impact such that it might merit publication in Nature. We therefore feel that the study would find a more suitable audience in another journal.
On July 1, Sørensen and his colleagues therefore challenged Science, another scientific journal to publish his article. Arguing for the publication of the article Sørensen wrote in an e-mail to editor Professor Holden Thorp:
“Now that Dr. Tedros Adhanom Ghebreyesus has indicated that WHO will pursue a long-overdue inquiry into the aetiology of the SARS-CoV-2 virus, which we welcome, we hope that you will support the very necessary debate that is now breaking in a second wave, following the publication of our vaccine. We are aware of significant responsible mainstream media interventions that are imminent. We are glad that this important question will now be addressed where it should be, in mainstream media and science journals, and not left to internet speculations, some of which have been both uninformed and therefore unhelpful, in our view.”
However, Sørensen was rebuffed also by Science the very next day. In an email to Sørensen, Professor Throp wrote that the article was unfit for publishing in Science, due to the fact that it criticizes work published in another journal.
Rejection from Science
Dr. Sorenson,
Thank you for your interest. We do not publish papers that are critiques of works in other journals, so we cannot consider something along these lines.
Holden
Holden Thorp
Editor-in-Chief
On the rejections from the scientific journals co-author Angus Dalgleish told Sky News: “I thought the whole point of a scientific journal was that you put forward some speculation and you opened it up to debate”, said Professor Dalgleish.
Agree or disagree with Dalgleish’s description of what a scientific journal does, the new round of rejections complicate the picture from the first round. A major part of the argument was accepted by a respectable journal -– the one that didn’t spell out the implications for the origin of the virus. The article that did spell it out, was now rejected twice, without peer review, and the second rejection on purely formal grounds that the authors vehemently contest, arguing that the paper in its current form is not first and foremost a critique of the Andersen et al. paper.
Minerva has asked both Nature and Science to elaborate on the reasons for why the articles by Sørensen and his co-authors have been rejected. Both Science and Nature have declined to comment on the specific rejection of the article as they view this information as confidential. However, Executive Director of the Science Press Package Meagan Phelan, replied that “Science receives upwards of 11,000 manuscripts per year, and the acceptance rate is 6%, so the vast majority of papers are rejected for one reason or another. Science’s acceptance rate for COVID-19-related submissions is even lower, at 4%. The journal continues to receive an exceptionally high volume of COVID-19-related submissions each week.”
At press time, the second article is still in search of a scientific journal willing to publish – or, at least, to subject it to peer review.
Correction: The name Quarterly Reviews of Biophysics has been changed to Quarterly Reviews of Biophysics Discovery
- admin
- Site Admin
- Posts: 36135
- Joined: Thu Aug 01, 2013 5:21 am
Re: U.S. government gave $3.7 million grant to Wuhan lab at
The evidence which suggests that this is no naturally evolved virus
by Aksel Fridstrøm
July 13, 2020 - 16:47
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
In the article “The Evidence which Suggests that This Is No Naturally Evolved Virus” by Sørensen, Dalgleish and Susrud, uploaded by Minerva today, Sørensen and his co-authors present several arguments for why they consider a non-natural origin for the novel coronavirus to be the most logical explanation on how the virus evolved.
It is observations on the spike protein of the Sars-Cov-2 virus that leads Sørensen and his co-authors to believe that the virus has originated in a laboratory.
“There are several factors that point towards this”, Sørensen told Minerva in an interview last week. “Firstly, this part of the virus is very stable; it mutates very little. That points to this virus as a fully developed, almost perfected virus for infecting humans.
“Secondly, this indicates that the structure of the virus cannot have evolved naturally. When we compare the novel coronavirus with the one that caused SARS, we see that there are altogether six inserts in this virus that stand out compared to other known SARS viruses”, he goes on explaining.
Sørensen says that several of these changes in the virus are unique, and that they do not exist in other known SARS coronaviruses.
“Four of these six changes have the property that they are suited to infect humans. This kind of aggregation of a type of property can be done simply in a laboratory, and helps to substantiate such an origin”, Sørensen argues.
Sørensen was also quoted saying that he is quite confident that the virus originated in a laboratory. “I think it’s more than 90 percent certain. It’s at least a far more probable explanation than it having developed this way in nature”, Sørensen said.
Sørensen also highlights other data than those related to the virus’ properties:
“The properties that we now see in the virus, we have yet to discover anywhere in nature. We know that these properties make the virus very infectious, so if it came from nature, there should also be many animals infected with this, but we have still not been able to trace the virus in nature”, Sørensen also elaborated in last week's interview.
Chimera
In the article Sørensen and his co-authors add additional context to how they believe the virus evolved through experiments in laboratories.
The authors do this by pointing to earlier and open articles by Chinese and American researchers, where the researchers demonstrate and discuss how they have manipulated new chimeraviruses into existence, with SARS-coronavirus as a starting point.
The word chimera derives from Greek mythology and refers to a fable being which was a third dragon, a third lion and a third goat.
Sørensen and his co-authors point to four different research projects where the chronology of the creation of a new chimeravirus is documented.
The first of these is the research project led by the world-famous Chinese scientist Dr. Zheng-Li Shi and her colleagues at Wuhan Virology Institute in 2008, which “successfully demonstrated technical capabilities to interchange RBD’s between bat SARS-like and human SARS viruses”.
These findings made the exchange of properties between SARS-coronavirus possible, Sørensen and his co-authors argue.
Furthermore, Sørensen’s article points to the fact that Wuhan’s Virology Institute again in 2010 took part in gain-of-function experiments with international collaborators, where SARS-coronavirus was provided with additional properties that increase the virus’s ability to infect humans: “In 2010 scientists from the ‘Special Viruses’ section of the Wuhan Institute of Virology were engaged in ‘gain of function’ experiments, jointly with international collaborators, to increase SARS-CoV infectiousness for humans. They used an HIV pseudovirus to express seven bat ACE2 receptors and compared their binding properties to human ACE2 receptors in order to pick the best for further optimizing a SARS-like coronavirus’s ability to bind to human cells”, they write.
Chimerica
In order to build further on this research, Shi and her colleagues sought help from American researchers.
“The Chinese originally lacked the technology and competence to grow and measure the development of the virus”, Sørensen argues.
However, according to Sørensen, Shi and her colleagues received help from the University of North Carolina at Chapel Hill in the US, where a human cell culture was made available for further research on SARS-coronavirus.
In the article, Sørensen and his co-authors write: “In 2015 scientists from the ‘Special Viruses’ section of the Wuhan Institute of Virology were engaged in ‘gain of function’ experiments jointly with a majority team from the University of North Carolina Chapel Hill. Together, they manipulated bat viruses to create a mouse adapted chimeric virus SHC014-MA15 which binds to and can proliferate on human upper airway cells (2B4 Calu-3 – a cell line contributed by Chapel Hill).”
Sørensen and his co-authors write that this work created “a chimeric virus with very high infectivity potential targeted to the human upper respiratory tract” and that what is being described is “in fact, precisely SARS-CoV-2 properties”, which is the virus that causes Covid-19.
Sørensen and his co-authors writes that “the 2015 authors were well aware that the chimeric virus which they had created was very dangerous because they discussed this fact”, but also explains that the creation of the chimeric virus of increased pathogenicity was not intentional. “In vivo experiments at Chapel Hill replicated the chimeric virus in mouse lung which showed significant pathogenesis which was the opposite of what the team had expected (“the creation of chimeric viruses like SHC014-MA15 was not expected to increase pathogenicity”).
The chimeric virus SHC014-MA15 that was created as a part of the collaboration between the Wuhan Institute of Virology and University of North Carolina at Chapel Hill is however according to an article published Shan-Lu Liu et al still different from the Sars-Cov-2 virus with a “divergence in the genetic sequence of this construct with the new SARS-CoV-2 (>5,000 nucleotides)”, Shan-Lu Liu writes.
Professor Kristian Andersen at the department of immunology and microbiology at Scripps Research again refers to the study conducted by Shan-Lu Liu when he writes that “the genetic data irrefutably show that SARS-CoV-2 is not derived from any previously used virus backbone”.
Sørensen and his co-authors is however not convinced that the divergence between the chimeric SHC014-MA15 virus created in 2015 and the novel coronavirus Sars-Cov-2 is enough to eliminate the possibility of Sars-Cov-2 being another chimeric virus that builds on this research.
To establish a connection between the chimeric viruses engineered by the Wuhan Institute of Virology and the Sars-CoV-2 virus Sørensen and his co-authors points to events back in 2016 when a new coronavirus started to infect pigs in China. This disease named SADS ended up killing approximately 25,000 piglets. The epidemic was investigated by the researcher at the Wuhan Institute of Virology, and tested against all known receptors used by coronaviruses, but none of these worked.
Sørensen and his co-authors therefore draws the conclusion that “SADS is a CoV infection utilising new tissue-specific binding domains”, and argues that the Sars-CoV-2 coronavirus has attained directly or indirectly properties from the SADS virus that makes it possible for the virus to use new receptors to attach to human cells in the upper airways.
Hence the evolution of the Sars-Cov-2 virus is summarized by Sørensen and his co-authors as follows: “In 2008, Dr Zheng-Li Si and WIV colleagues successfully demonstrated technical capabilities to interchange RBD’s between bat SARS-like and human SARS viruses. Building upon this, the 2010 work (Hou et al, 2010) perfected the ability to express receptors on human cells. On these foundations, the central Gain of Function work that underpins the functionalities of SARS-CoV-2 took place, carrying the WIV spike and plasmid materials to bond successfully to a UNC Chapel Hill human epithelial cell-line.
This work produced a highly infectious chimeric virus optimised to the human upper respiratory tract. In convergent support of this hypothesis, both Lu and Jia have now, in January and April 2020, shown that SARS-CoV-2 has a bat SARS-like backbone but is carrying an RBD from a human SARS and Zhan et al have, like us, noted unusual adaption to humans from the first isolate.
In the 2015 Chapel Hill work it was only ACE2 receptors that were discussed. However, in 2018 Zhou P. et al demonstrated capabilities to clone other receptors like APN and DPP4 and to test and compare these against the (intestine) tissue specific SADS-CoV identified. Then, in the 2019-20 Covid-19 pandemic, profuse symptoms indicating compromise of the bitter/sweet receptors are reported. Taken all together, this implies that by employing insights gained after 2015, as just deduced, a further optimization of the 2015 chimeric virus for additional binding to receptors/co-receptors such as bitter/sweet specific upper airway epithelia receptors occurred. That would help to explain the otherwise puzzling high infectivity and pathology associated with SARS-CoV-2 and hence also help to explain the social epidemiology of its spread”.
Thinking the unthinkable
The release of a potential pandemic pathogen from a laboratory is by many viewed as a too catastrophic event to even be considered.
However many scientists now argue that it has already happened. In 1977 the H1N1 influenza virus caused an epidemic in the Soviet Union and China. This rapidly spreading epidemic was almost entirely restricted to persons younger than 26 years of age. A common explanation for this oddity is that the epidemic was caused by a frozen version of the virus that circulated in the 1950s, and in some way or another escaped laboratory containment.
It is also a known fact that the SARS-coronavirus that caused the SARS epidemic in 2003 has escaped laboratories in South-East Asia on at least four occasions. In 2003 WHO issued a statement calling for increased safety measures in laboratories that handle SARS-coronaviruses highlighting the risk of lab accidents: “The possibility that a SARS outbreak could occur following a laboratory accident is a risk of considerable importance, given the relatively large number of laboratories currently conducting research using the SARS-CoV or retaining specimens from SARS patients. These laboratories currently represent the greatest threat for renewed SARS-CoV transmission through accidental exposure associated with breaches in laboratory biosafety”, the WHO stated.
The laboratory operated by the Wuhan Institute of Virology was however built to adhere to far stricter security protocols than the ones criticized by the WHO back in 2003, and became the first Chinese laboratory that operated at the highest level of biosafety (level 4), when it was inaugurated in 2015.
At the Wuhan Institute of Virology lead scientist Dr. Shi Zhengli ferociously denies any possibility that the Sars-cov-2 virus is a lab escapee. In a rare interview with the Scientific American, Zhengli explains that “she frantically went through her own lab’s records from the past few years to check for any mishandling of experimental materials, especially during disposal. Shi breathed a sigh of relief when the results came back: none of the sequences matched those of the viruses her team had sampled from bat caves”.
“That really took a load off my mind”, she is quoted saying.
Dr. Zhengli has also published the genomic sequence of a new virus that her team had sampled in an abandoned mine shaft seven years ago that shares an overall genome sequence identity of 96.2% with the Sars-Cov-2 virus, the closest match so far according to her article.
However, concerns about the operational safety at the Wuhan Institute of Virology persist. In an article written by Josh Rogin at The Washington Post, Rogin refers to a leak embassy cable sent from the American Embassy in Beijing that raised on concerns about the safety routines at the Wuhan laboratory
“During interactions with scientists at the WIV laboratory, they noted the new lab has a serious shortage of appropriately trained technicians and investigators needed to safely operate this high-containment laboratory”, states the Jan. 19, 2018, cable, which was drafted by two officials from the embassy’s environment, science and health sections who met with the scientists at the Wuhan Institute of Virology.
WHO to investigate
On the 18-19th of May the WHO conducted its 73rd general assembly, and passed a resolution that “requests the Director-General, working with other organizations and countries, to identify the zoonotic source of the virus and the route of introduction to the human population”.
Sørensen and his co-authors argue that this investigation should not be limited to examining the origin of the Sars-Cov-2 virus solely based on the assumption of a zoonotic source.
The WHO investigation commenced on the 7th of July.
by Aksel Fridstrøm
July 13, 2020 - 16:47
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
YOU ARE REQUIRED TO READ THE COPYRIGHT NOTICE AT THIS LINK BEFORE YOU READ THE FOLLOWING WORK, THAT IS AVAILABLE SOLELY FOR PRIVATE STUDY, SCHOLARSHIP OR RESEARCH PURSUANT TO 17 U.S.C. SECTION 107 AND 108. IN THE EVENT THAT THE LIBRARY DETERMINES THAT UNLAWFUL COPYING OF THIS WORK HAS OCCURRED, THE LIBRARY HAS THE RIGHT TO BLOCK THE I.P. ADDRESS AT WHICH THE UNLAWFUL COPYING APPEARED TO HAVE OCCURRED. THANK YOU FOR RESPECTING THE RIGHTS OF COPYRIGHT OWNERS.
In the article “The Evidence which Suggests that This Is No Naturally Evolved Virus” by Sørensen, Dalgleish and Susrud, uploaded by Minerva today, Sørensen and his co-authors present several arguments for why they consider a non-natural origin for the novel coronavirus to be the most logical explanation on how the virus evolved.
It is observations on the spike protein of the Sars-Cov-2 virus that leads Sørensen and his co-authors to believe that the virus has originated in a laboratory.
FACTBOX – Spike Protein
A spike protein is a part of the virus attached to the surface of the virus. The spike protein is used by the virus when it enters cells, enabling it to stick in humans. The properties of the spike determine which receptors a virus can utilise and thus which cells the virus can enter to create illness.
“There are several factors that point towards this”, Sørensen told Minerva in an interview last week. “Firstly, this part of the virus is very stable; it mutates very little. That points to this virus as a fully developed, almost perfected virus for infecting humans.
“Secondly, this indicates that the structure of the virus cannot have evolved naturally. When we compare the novel coronavirus with the one that caused SARS, we see that there are altogether six inserts in this virus that stand out compared to other known SARS viruses”, he goes on explaining.
Sørensen says that several of these changes in the virus are unique, and that they do not exist in other known SARS coronaviruses.
“Four of these six changes have the property that they are suited to infect humans. This kind of aggregation of a type of property can be done simply in a laboratory, and helps to substantiate such an origin”, Sørensen argues.
Sørensen was also quoted saying that he is quite confident that the virus originated in a laboratory. “I think it’s more than 90 percent certain. It’s at least a far more probable explanation than it having developed this way in nature”, Sørensen said.
Sørensen also highlights other data than those related to the virus’ properties:
“The properties that we now see in the virus, we have yet to discover anywhere in nature. We know that these properties make the virus very infectious, so if it came from nature, there should also be many animals infected with this, but we have still not been able to trace the virus in nature”, Sørensen also elaborated in last week's interview.
Chimera
In the article Sørensen and his co-authors add additional context to how they believe the virus evolved through experiments in laboratories.
The authors do this by pointing to earlier and open articles by Chinese and American researchers, where the researchers demonstrate and discuss how they have manipulated new chimeraviruses into existence, with SARS-coronavirus as a starting point.
FACTBOX – Chimeravirus
Chimeravirus is defined by U.S. Department of Agriculture's Animal and Plant Health Inspection Service as a hybrid micro-organism which is created by combining fragments from two or more micro-organisms where at least one of the two fragments contain the necessary genes for replication.
The word chimera derives from Greek mythology and refers to a fable being which was a third dragon, a third lion and a third goat.
Sørensen and his co-authors point to four different research projects where the chronology of the creation of a new chimeravirus is documented.
The first of these is the research project led by the world-famous Chinese scientist Dr. Zheng-Li Shi and her colleagues at Wuhan Virology Institute in 2008, which “successfully demonstrated technical capabilities to interchange RBD’s between bat SARS-like and human SARS viruses”.
These findings made the exchange of properties between SARS-coronavirus possible, Sørensen and his co-authors argue.
Furthermore, Sørensen’s article points to the fact that Wuhan’s Virology Institute again in 2010 took part in gain-of-function experiments with international collaborators, where SARS-coronavirus was provided with additional properties that increase the virus’s ability to infect humans: “In 2010 scientists from the ‘Special Viruses’ section of the Wuhan Institute of Virology were engaged in ‘gain of function’ experiments, jointly with international collaborators, to increase SARS-CoV infectiousness for humans. They used an HIV pseudovirus to express seven bat ACE2 receptors and compared their binding properties to human ACE2 receptors in order to pick the best for further optimizing a SARS-like coronavirus’s ability to bind to human cells”, they write.
FACTBOX – gain-of-function-research
Gain-of-function studies are according to the US Department Health and Human Services research which involves increasing the capacity of a pathogen to cause illness. The method is controversial because it can also risk new viruses leaking out of laboratories and into the population.
In the period 2014 to 2018 this type of research was prohibited in the US, but in December 2017 American authorities announced that these kinds of studies would again be allowed.
Chimerica
In order to build further on this research, Shi and her colleagues sought help from American researchers.
“The Chinese originally lacked the technology and competence to grow and measure the development of the virus”, Sørensen argues.
However, according to Sørensen, Shi and her colleagues received help from the University of North Carolina at Chapel Hill in the US, where a human cell culture was made available for further research on SARS-coronavirus.
In the article, Sørensen and his co-authors write: “In 2015 scientists from the ‘Special Viruses’ section of the Wuhan Institute of Virology were engaged in ‘gain of function’ experiments jointly with a majority team from the University of North Carolina Chapel Hill. Together, they manipulated bat viruses to create a mouse adapted chimeric virus SHC014-MA15 which binds to and can proliferate on human upper airway cells (2B4 Calu-3 – a cell line contributed by Chapel Hill).”
Sørensen and his co-authors write that this work created “a chimeric virus with very high infectivity potential targeted to the human upper respiratory tract” and that what is being described is “in fact, precisely SARS-CoV-2 properties”, which is the virus that causes Covid-19.
Sørensen and his co-authors writes that “the 2015 authors were well aware that the chimeric virus which they had created was very dangerous because they discussed this fact”, but also explains that the creation of the chimeric virus of increased pathogenicity was not intentional. “In vivo experiments at Chapel Hill replicated the chimeric virus in mouse lung which showed significant pathogenesis which was the opposite of what the team had expected (“the creation of chimeric viruses like SHC014-MA15 was not expected to increase pathogenicity”).
The chimeric virus SHC014-MA15 that was created as a part of the collaboration between the Wuhan Institute of Virology and University of North Carolina at Chapel Hill is however according to an article published Shan-Lu Liu et al still different from the Sars-Cov-2 virus with a “divergence in the genetic sequence of this construct with the new SARS-CoV-2 (>5,000 nucleotides)”, Shan-Lu Liu writes.
Professor Kristian Andersen at the department of immunology and microbiology at Scripps Research again refers to the study conducted by Shan-Lu Liu when he writes that “the genetic data irrefutably show that SARS-CoV-2 is not derived from any previously used virus backbone”.
Sørensen and his co-authors is however not convinced that the divergence between the chimeric SHC014-MA15 virus created in 2015 and the novel coronavirus Sars-Cov-2 is enough to eliminate the possibility of Sars-Cov-2 being another chimeric virus that builds on this research.
To establish a connection between the chimeric viruses engineered by the Wuhan Institute of Virology and the Sars-CoV-2 virus Sørensen and his co-authors points to events back in 2016 when a new coronavirus started to infect pigs in China. This disease named SADS ended up killing approximately 25,000 piglets. The epidemic was investigated by the researcher at the Wuhan Institute of Virology, and tested against all known receptors used by coronaviruses, but none of these worked.
Sørensen and his co-authors therefore draws the conclusion that “SADS is a CoV infection utilising new tissue-specific binding domains”, and argues that the Sars-CoV-2 coronavirus has attained directly or indirectly properties from the SADS virus that makes it possible for the virus to use new receptors to attach to human cells in the upper airways.
Hence the evolution of the Sars-Cov-2 virus is summarized by Sørensen and his co-authors as follows: “In 2008, Dr Zheng-Li Si and WIV colleagues successfully demonstrated technical capabilities to interchange RBD’s between bat SARS-like and human SARS viruses. Building upon this, the 2010 work (Hou et al, 2010) perfected the ability to express receptors on human cells. On these foundations, the central Gain of Function work that underpins the functionalities of SARS-CoV-2 took place, carrying the WIV spike and plasmid materials to bond successfully to a UNC Chapel Hill human epithelial cell-line.
This work produced a highly infectious chimeric virus optimised to the human upper respiratory tract. In convergent support of this hypothesis, both Lu and Jia have now, in January and April 2020, shown that SARS-CoV-2 has a bat SARS-like backbone but is carrying an RBD from a human SARS and Zhan et al have, like us, noted unusual adaption to humans from the first isolate.
In the 2015 Chapel Hill work it was only ACE2 receptors that were discussed. However, in 2018 Zhou P. et al demonstrated capabilities to clone other receptors like APN and DPP4 and to test and compare these against the (intestine) tissue specific SADS-CoV identified. Then, in the 2019-20 Covid-19 pandemic, profuse symptoms indicating compromise of the bitter/sweet receptors are reported. Taken all together, this implies that by employing insights gained after 2015, as just deduced, a further optimization of the 2015 chimeric virus for additional binding to receptors/co-receptors such as bitter/sweet specific upper airway epithelia receptors occurred. That would help to explain the otherwise puzzling high infectivity and pathology associated with SARS-CoV-2 and hence also help to explain the social epidemiology of its spread”.
Thinking the unthinkable
The release of a potential pandemic pathogen from a laboratory is by many viewed as a too catastrophic event to even be considered.
However many scientists now argue that it has already happened. In 1977 the H1N1 influenza virus caused an epidemic in the Soviet Union and China. This rapidly spreading epidemic was almost entirely restricted to persons younger than 26 years of age. A common explanation for this oddity is that the epidemic was caused by a frozen version of the virus that circulated in the 1950s, and in some way or another escaped laboratory containment.
It is also a known fact that the SARS-coronavirus that caused the SARS epidemic in 2003 has escaped laboratories in South-East Asia on at least four occasions. In 2003 WHO issued a statement calling for increased safety measures in laboratories that handle SARS-coronaviruses highlighting the risk of lab accidents: “The possibility that a SARS outbreak could occur following a laboratory accident is a risk of considerable importance, given the relatively large number of laboratories currently conducting research using the SARS-CoV or retaining specimens from SARS patients. These laboratories currently represent the greatest threat for renewed SARS-CoV transmission through accidental exposure associated with breaches in laboratory biosafety”, the WHO stated.
The laboratory operated by the Wuhan Institute of Virology was however built to adhere to far stricter security protocols than the ones criticized by the WHO back in 2003, and became the first Chinese laboratory that operated at the highest level of biosafety (level 4), when it was inaugurated in 2015.
At the Wuhan Institute of Virology lead scientist Dr. Shi Zhengli ferociously denies any possibility that the Sars-cov-2 virus is a lab escapee. In a rare interview with the Scientific American, Zhengli explains that “she frantically went through her own lab’s records from the past few years to check for any mishandling of experimental materials, especially during disposal. Shi breathed a sigh of relief when the results came back: none of the sequences matched those of the viruses her team had sampled from bat caves”.
“That really took a load off my mind”, she is quoted saying.
Dr. Zhengli has also published the genomic sequence of a new virus that her team had sampled in an abandoned mine shaft seven years ago that shares an overall genome sequence identity of 96.2% with the Sars-Cov-2 virus, the closest match so far according to her article.
The hypothesis that SARS-CoV-2 evolved in the Mojiang miner’s lungs potentially resolves many scientific questions about the origin of the pandemic. But it raises others having to do with why this information has not come to light hitherto. The most obvious of these concern the actions of the Shi lab at the WIV.
Why did the Shi lab not acknowledge the miners’ deaths in any paper describing samples taken from the mine (Ge et al., 2016 and P. Zhou et al., 2020)? Why in the title of the Ge at al. 2016 paper did the Shi lab call it an “abandoned” mine? When they published the sequence of RaTG13 in Feb. 2020, why did the Shi lab provide a new name (RaTG13) for BtCoV/4991 when they had by then cited BtCoV/4991 twice in publications and once in a genome sequence database and when their sequences were from the same sample and 100% identical (P. Zhou et al., 2020)? If it was just a name change, why no acknowledgement of this in their 2020 paper describing RaTG13 (Bengston, 2020)? These strange and unscientific actions have obscured the origins of the closest viral relatives of SARS-CoV-2, viruses that are suspected to have caused a COVID-like illness in 2012 and which may be key to understanding not just the origin of the COVID-19 pandemic but the future behaviour of SARS-CoV-2.
These are not the only questionable actions associated with the provenance of samples from the mine. There were five scientific publications that very early in the pandemic reported whole genome sequences for SARS-CoV-2 (Chan et al., 2020; Chen et al., 2020; Wu et al., 2020; P. Zhou et al., 2020; Zhu et al., 2020). Despite three of them having experienced viral evolutionary biologists as authors (George Gao, Zheng-li Shi and Edward Holmes) only one of these (Chen et al., 2020) succeeded in identifying the most closely related viral sequence by far: BtCoV/4991 a viral sequence in the possession of the Shi lab at the WIV that differed from SARS-CoV-2 by just 5 nucleotides.
-- A Proposed Origin for SARS-CoV-2 and the COVID-19 Pandemic, by Jonathan Latham, PhD and Allison Wilson, PhD
However, concerns about the operational safety at the Wuhan Institute of Virology persist. In an article written by Josh Rogin at The Washington Post, Rogin refers to a leak embassy cable sent from the American Embassy in Beijing that raised on concerns about the safety routines at the Wuhan laboratory
“During interactions with scientists at the WIV laboratory, they noted the new lab has a serious shortage of appropriately trained technicians and investigators needed to safely operate this high-containment laboratory”, states the Jan. 19, 2018, cable, which was drafted by two officials from the embassy’s environment, science and health sections who met with the scientists at the Wuhan Institute of Virology.
Two years before the novel coronavirus pandemic upended the world, U.S. Embassy officials visited a Chinese research facility in the city of Wuhan several times and sent two official warnings back to Washington about inadequate safety at the lab, which was conducting risky studies on coronaviruses from bats. The cables have fueled discussions inside the U.S. government about whether this or another Wuhan lab was the source of the virus — even though conclusive proof has yet to emerge.
In January 2018, the U.S. Embassy in Beijing took the unusual step of repeatedly sending U.S. science diplomats to the Wuhan Institute of Virology (WIV), which had in 2015 become China’s first laboratory to achieve the highest level of international bioresearch safety (known as BSL-4). WIV issued a news release in English about the last of these visits, which occurred on March 27, 2018. The U.S. delegation was led by Jamison Fouss, the consul general in Wuhan, and Rick Switzer, the embassy’s counselor of environment, science, technology and health. Last week, WIV erased that statement from its website, though it remains archived on the Internet.
What the U.S. officials learned during their visits concerned them so much that they dispatched two diplomatic cables categorized as Sensitive But Unclassified back to Washington. The cables warned about safety and management weaknesses at the WIV lab and proposed more attention and help. The first cable, which I obtained, also warns that the lab’s work on bat coronaviruses and their potential human transmission represented a risk of a new SARS-like pandemic.
“During interactions with scientists at the WIV laboratory, they noted the new lab has a serious shortage of appropriately trained technicians and investigators needed to safely operate this high-containment laboratory,” states the Jan. 19, 2018, cable, which was drafted by two officials from the embassy’s environment, science and health sections who met with the WIV scientists. (The State Department declined to comment on this and other details of the story.)
The Chinese researchers at WIV were receiving assistance from the Galveston National Laboratory at the University of Texas Medical Branch and other U.S. organizations, but the Chinese requested additional help. The cables argued that the United States should give the Wuhan lab further support, mainly because its research on bat coronaviruses was important but also dangerous.
As the cable noted, the U.S. visitors met with Shi Zhengli, the head of the research project, who had been publishing studies related to bat coronaviruses for many years. In November 2017, just before the U.S. officials’ visit, Shi’s team had published research showing that horseshoe bats they had collected from a cave in Yunnan province were very likely from the same bat population that spawned the SARS coronavirus in 2003.
“Most importantly,” the cable states, “the researchers also showed that various SARS-like coronaviruses can interact with ACE2, the human receptor identified for SARS-coronavirus. This finding strongly suggests that SARS-like coronaviruses from bats can be transmitted to humans to cause SARS-like diseases. From a public health perspective, this makes the continued surveillance of SARS-like coronaviruses in bats and study of the animal-human interface critical to future emerging coronavirus outbreak prediction and prevention.”
The research was designed to prevent the next SARS-like pandemic by anticipating how it might emerge. But even in 2015, other scientists questioned whether Shi’s team was taking unnecessary risks. In October 2014, the U.S. government had imposed a moratorium on funding of any research that makes a virus more deadly or contagious, known as “gain-of-function” experiments.
As many have pointed out, there is no evidence that the virus now plaguing the world was engineered; scientists largely agree it came from animals. But that is not the same as saying it didn’t come from the lab, which spent years testing bat coronaviruses in animals, said Xiao Qiang, a research scientist at the School of Information at the University of California at Berkeley.
“The cable tells us that there have long been concerns about the possibility of the threat to public health that came from this lab’s research, if it was not being adequately conducted and protected,” he said.
There are similar concerns about the nearby Wuhan Center for Disease Control and Prevention lab, which operates at biosecurity level 2, a level significantly less secure than the level-4 standard claimed by the Wuhan Insititute of Virology lab, Xiao said. That’s important because the Chinese government still refuses to answer basic questions about the origin of the novel coronavirus while suppressing any attempts to examine whether either lab was involved.
Sources familiar with the cables said they were meant to sound an alarm about the grave safety concerns at the WIV lab, especially regarding its work with bat coronaviruses. The embassy officials were calling for more U.S. attention to this lab and more support for it, to help it fix its problems.
“The cable was a warning shot,” one U.S. official said. “They were begging people to pay attention to what was going on.”
No extra assistance to the labs was provided by the U.S. government in response to these cables. The cables began to circulate again inside the administration over the past two months as officials debated whether the lab could be the origin of the pandemic and what the implications would be for the U.S. pandemic response and relations with China.
Inside the Trump administration, many national security officials have long suspected either the WIV or the Wuhan Center for Disease Control and Prevention lab was the source of the novel coronavirus outbreak. According to the New York Times, the intelligence community has provided no evidence to confirm this. But one senior administration official told me that the cables provide one more piece of evidence to support the possibility that the pandemic is the result of a lab accident in Wuhan.
“The idea that it was just a totally natural occurrence is circumstantial. The evidence it leaked from the lab is circumstantial. Right now, the ledger on the side of it leaking from the lab is packed with bullet points and there’s almost nothing on the other side,” the official said.
As my colleague David Ignatius noted, the Chinese government’s original story — that the virus emerged from a seafood market in Wuhan — is shaky. Research by Chinese experts published in the Lancet in January showed the first known patient, identified on Dec. 1, had no connection to the market, nor did more than one-third of the cases in the first large cluster. Also, the market didn’t sell bats.
Shi and other WIV researchers have categorically denied this lab was the origin for the novel coronavirus. On Feb. 3, her team was the first to publicly report the virus known as 2019-nCoV was a bat-derived coronavirus.
The Chinese government, meanwhile, has put a total lockdown on information related to the virus origins. Beijing has yet to provide U.S. experts with samples of the novel coronavirus collected from the earliest cases. The Shanghai lab that published the novel coronavirus genome on Jan. 11 was quickly shut down by authorities for “rectification.” Several of the doctors and journalists who reported on the spread early on have disappeared.
On Feb. 14, Chinese President Xi Jinping called for a new biosecurity law to be accelerated. On Wednesday, CNN reported the Chinese government has placed severe restrictions requiring approval before any research institution publishes anything on the origin of the novel coronavirus.
The origin story is not just about blame. It’s crucial to understanding how the novel coronavirus pandemic started because that informs how to prevent the next one. The Chinese government must be transparent and answer the questions about the Wuhan labs because they are vital to our scientific understanding of the virus, said Xiao.
We don’t know whether the novel coronavirus originated in the Wuhan lab, but the cable pointed to the danger there and increases the impetus to find out, he said.
“I don’t think it’s a conspiracy theory. I think it’s a legitimate question that needs to be investigated and answered,” he said. “To understand exactly how this originated is critical knowledge for preventing this from happening in the future.”
-- State Department cables warned of safety issues at Wuhan lab studying bat coronaviruses, by Josh Rogin, The Washington Post, April 14, 2020
WHO to investigate
On the 18-19th of May the WHO conducted its 73rd general assembly, and passed a resolution that “requests the Director-General, working with other organizations and countries, to identify the zoonotic source of the virus and the route of introduction to the human population”.
Sørensen and his co-authors argue that this investigation should not be limited to examining the origin of the Sars-Cov-2 virus solely based on the assumption of a zoonotic source.
The WHO investigation commenced on the 7th of July.
- admin
- Site Admin
- Posts: 36135
- Joined: Thu Aug 01, 2013 5:21 am
Re: U.S. government gave $3.7 million grant to Wuhan lab at
Laboratory Escapes and “Self-fulfilling prophecy” Epidemics
by Martin Furmanski MD
Scientist’s Working Group on Chemical and Biologic Weapons
Center for Arms Control and Nonproliferation
February 17, 2014
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
Introduction
The danger to world or regional public health from the escape from microbiology laboratories of pathogens capable of causing pandemics, or Potentially Pandemic Pathogens (PPPs) has been the subject of considerable discussion1,2,3,4 including mathematical modeling of the probability and impact of such escapes5. The risk of such releases has generally been determined from estimates of laboratory infections that are often incomplete, except for the recent 2013 Centers for Disease Control (CDC) report6, which is a significant source of recent data on escapes from undetected and unreported laboratory-acquired infections (LAIs).
This paper presents an historical review of outbreaks of PPPs or similarly transmissible pathogens that occurred from presumably well-funded and supervised nationally supported laboratories. It should be emphasized that these examples are only the “tip of the iceberg” because they represent laboratory accidents that have actually caused illness outside of the laboratory in the general public environment. The list of laboratory workers who have contracted potentially contagious infections in microbiology labs but did not start community outbreaks is much, much longer. The examples here are not “near misses;” these escapes caused real-world outbreaks.
Methods of pathogen identification
Modern genetic analysis allows pathogens to be identified, and given a sufficient catalog of isolates of the same pathogen, it is possible to determine if two specimens are identical or very closely related. Because all pathogens that are circulating in the environment show genetic changes over time, one can date the time the pathogen circulated. For instance, for 20th century human and swine influenza viruses beginning in the 1930s, one can generally place a virus to a particular year. With modern rapid genomic analysis outbreaks can be traced with considerable accuracy: for instance the 2009 pandemic pH1N1 influenza outbreak has been analyzed with confidence limits of branchpoints in its first wave defined within days or weeks, and individual transmission chains can be identified7.
Example #1: British smallpox escapes, 1966, 1972, 1978
The WHO’s successful effort to eradicate natural transmission of smallpox in the 1970s highlighted the risk that virology laboratories posed as a source of epidemics. This was clearly demonstrated in the United Kingdom, where from 1963-1978 only 4 cases of smallpox (with no deaths) were reported from smallpox endemic areas, while during the same period at least 80 cases and 3 deaths were the result of three separate escapes of the smallpox virus from two different accredited smallpox laboratories.8 Much of the current policy and practice in biosafety and biocontainment of dangerous pathogens can be traced to the political and professional reaction to these outbreaks.
The UK became a sensitive test system for smallpox laboratory escapes because it ended compulsory smallpox vaccination in 1946. Public sentiment in the UK had always included significant resistance to and apathy towards vaccination, and so by the mid 1960s and through the 1970s a large proportion of children and young adults had never been vaccinated, and many older persons were never re-vaccinated after initial childhood or military vaccinations. Thus the protective herd immunity in the general public, which earlier rendered impotent any laboratory escapes, disappeared. At the same time, the considerable volume of travel and immigration from smallpox endemic areas of Africa and the Indian subcontinent meant that surveillance for imported smallpox cases was required. UK maintained several smallpox laboratories at medical schools for both research and to support clinical diagnosis.
The first laboratory outbreak to be recognized began in March 1972, in a 23 year old laboratory assistant at the London School of Hygiene and Tropical Medicine, who had observed harvesting of live smallpox virus from eggs. This had been done on an open bench, as was routine, the laboratory having no isolation cabinets at that time. Before she was placed in isolation, she infected two visitors to a patient in an adjacent bed, both of whom died. They in turn infected a nurse, who survived9.
The recognition of this laboratory escape resulted in several investigations, which led to the establishment of guidelines for laboratories handling smallpox and other dangerous pathogens. These recommendations included handling dangerous pathogens in biological safety cabinets only in certain dedicated rooms by specifically trained and designated personnel. Also guidelines were issued for isolation with dedicated gowns and gloves, and the establishment of proper ventilation facilities to maintain negative pressure in these rooms and cabinets. These recommendations are the direct precursors to the current Biosafety Laboratory (BSL) level protocols.
By 1977 the natural chain of smallpox transmission had been interrupted, and the WHO was in the process of reducing the number of laboratories holding smallpox virus. In August of 1978 a 40 year old medical photographer at Birmingham Medical School developed smallpox, and died. She infected her mother, who survived. She worked in a studio and darkroom that was immediately above the smallpox laboratory at Birmingham Medical School. Investigation revealed that although the long established laboratory had been inspected and approved to handle smallpox virus, it did not have sufficient facilities to meet the new biocontainment requirements, and was scheduled to be decommissioned at the end of 1978. Moreover, work on smallpox had accelerated substantially in order to complete existing projects before the closing, and work with smallpox was performed by laboratory personnel who did not receive appropriate training and supervision, and appropriate isolation practices were frequently violated. The most likely route of exposure of the Medical Photographer was by transport of infectious aerosols generated by a centrifuge through building ventilation ducts that were improperly sealed and allowed aerosols to be delivered to one of the Photographer’s working spaces. Laboratory notebooks and the photographer’s work logs indicated that the strain infecting the photographer was handled in the laboratory on the same days that the photographer worked in the potentially contaminated workspace, on dates consistent with the photographer’s calculated exposure date. Dr Henry Bedson, a world renowned smallpox investigator who was responsible for the Birmingham laboratory, committed suicide as a result of the outbreak (Shooter 1980).
The 1978 investigation re-examined a 1966 smallpox outbreak, which in retrospect was strikingly similar to the 1978 outbreak. The earliest case identified in 1966 was in a medical photographer who worked at Birmingham Medical School in the same facility as the 1978 case. This outbreak was caused by a low-virulence strain of smallpox (variola minor), and it caused at least 72 cases of smallpox from February to August 1966, spread through the midlands of Britain, and Wales. The vast majority of cases were in unvaccinated children or young adults. There were no deaths. Retrospective review again revealed variola minor had been manipulated in the smallpox laboratory at a time appropriate to cause the infection in the photographer working a floor above.
Example #2: The “re-emergence” of H1N1 human influenza in 1977.
Human influenza H1N1 viruses appeared with the 1918 pandemic, and persisted, slowing accumulating small changes in its genome (with a major change in 1947), until the H2N2 “Asian” flu appeared in 1957, causing a worldwide pandemic. H1N1 influenza virus then apparently became extinct, and was not isolated for 20 years. In 1969 the “Hong Kong” H3N2 virus replaced the H2N2 virus, and is still circulating.
In September 1977 an H1N1 influenza virus was isolated from human infections in the Far East region of the Soviet Union, and in early 1978 the Chinese reported they had isolated H1N1 virus in May of 1977 in northeast China adjacent to the Soviet outbreak1011. Using the early genetic tools available at the time, the 1977 H1N1 virus was found to be closely related to H1N1 human influenza viruses circulating in 1949-1950, but not to those circulating earlier or later12, 13.
The 1977 H1N1 flu virus rapidly spread worldwide, in a pandemic that was restricted largely to people under ~ 21 years of age. Older persons had been exposed to related H1N1 viruses prior to 1957, and carried substantial immunity. Mercifully, the “re-emergent” H1N1 virus was not very virulent. Although illness was widespread, affecting 20-70% of those under 20 years of age in school or military camp outbreaks in the first year14, deaths were few. Many asymptomatic infections were detected by serology (Kung 1978).
The appearance of this “time-traveling throwback” puzzled virologists, because no similar examples had previously been identified in influenza or other similar viruses. Initially escape of a virus kept in storage from c1950 from a virology lab was discussed, but such a laboratory accident was denied by Chinese and Soviet virologists (Kung 1978, Beveridge 1978). Western virologists quietly let the matter of a laboratory escape origin for the 1977 H1N1 virus drop from discussion, out of an abundance of scientific caution, and also out of an eagerness not to offend the Russian and Chinese scientists, whose early gestures of cooperation in worldwide influenza surveillance system were very important to foster, because such cooperation would allow tracking influenza globally.
Discussions of the origins of the 1977 H1N1 gave rise to hypotheses of natural “biological stasis” or viral latency in an undefined animal. Experimental investigations of possible transmission of human H1N1 viruses in avians were pursued, but with minimal success and no demonstration of persistent avian transmission15, nor were human viruses identified in avians in very extensive subsequent surveys. The ambiguous term “frozen evolution” was coined, allowing for the freezing to be biologically functional, metaphorical, refrigerative, or natural.
A 2006 paper16 claimed to have isolated H1N1 influenza virus RNA from ice and meltwater from Siberian lakes that were frequented by migratory birds. Since migratory birds naturally carry and shed a wide variety of influenza viruses, and since year to year variations in the amount of thawing of lake ice might allow influenza viruses shed from the migratory birds to remain physically frozen for a number of years, the paper stated this might be the mechanism for the re-emergence of the 1977 H1N1 flu. It emphasized the 1977 H1N1 link because the RNA sequences it reported isolating from the lakes were closely related to sequences of three H1N1 reference viruses that it characterized as being of avian origin that circulated in the late 1960s.
Problems soon arose with this paper, however. The authors issued a correction17 in 2007 indicating the H1N1 reference strains originally characterized as avian and from the 1960s were in fact of human origin, and dated from the 1930s. A paper highly critical of the 2006 Siberian Lake paper was published in 2008 18, presenting strong evidence that the reported isolation of influenza RNA from nature was the result of contamination in the laboratory by the standard reference strain of human H1N1 virus (isolated in 1933) that was used as a positive control in that laboratory.
Presently, with detailed sophisticated genomic analysis available, and with 32 years of circulation of the 1977 H1N1 virus available for study, no evidence of natural genomic stasis has been identified. It has become clear that its appearance in 1977 was almost certainly due to escape from a virology lab of a virus sample that had been frozen since c1950. Only since 2008 have virologists actually begun to make the suggestion of a probable laboratory release in scientific papers: “The reemergence in 1977 is unexplained and probably represents reintroduction to humans from a laboratory source19,” and “…little A/H1N1 evolution is evident over the twenty-year period of the virus’s global disappearance, supporting earlier suggestions that this subtype was most likely accidentally reintroduced into human circulation from a laboratory environment20.” It should be noted that this paper calculates the 1977 H1N1 virus had been circulating for 1 year before it was reported, so that geographic origin cannot be stated with certainty.
Only since 2009-2010 did major papers begin to state directly the 1977 emergence of H1N1 influenza was a laboratory related release: “The most famous case of a released laboratory strain is the re-emergent H1N1 influenza A virus which was first observed in China in May of 1977 and in Russia shortly thereafter21.” The paper made this statement in part because the continued “agnostic” approach to the 1977 re-emergence introduced unacceptable errors in calculating the genomic divergence dates for influenza virus strains.
Public awareness of the 1977 H1N1 pandemic and its likely laboratory origins has been virtually absent. Virologists and public health officials with the appropriate sophistication were quickly aware that a laboratory release was the most likely origin, but they were content not to publicize this, aware that such embarrassing allegations would likely end the then nascent cooperation of Russian and Chinese virologists, which was vital to worldwide influenza surveillance. An abundance of caution in making such suggestions was also in their own self-interest. The 1976 “swine flu” alarm and subsequent immunization program that proved to be unneeded caused 532 cases of Guillain-Barre syndrome and 32 deaths. It was widely considered a misadventure, and had severely damaged the public and political credibility of the virology and public health communities. An acknowledgement of a pandemic originating from their laboratories would have only worsened it. The most plausible reason for a Chinese or Russian laboratory to thaw out and begin growing a c1950 H1N1 virus in 1976-77 was as a response to the US 1976 “swine flu” program, which resulted in a program to immunize the entire US population against H1N1 influenza virus. It was clearly a rational response for other countries with virology capabilities to explore making their own H1N1 vaccines. Thawing available frozen stocks of virus was necessary, because H1N1 was no longer circulating. Modern commentators have begun to articulate this connection between the 1976 Swine flu immunization program and the 1977 H1N1 re-emergence:
The speculation that the 1977 release may have been related to H1N1 vaccine research is supported by the observation that in the initial outbreaks in China, nine of the ten viral isolates expressed “temperature sensitivity” (Kung 1978). Temperature sensitivity normally an uncommon trait, but one that was in the 1970s (and still is) a fundamental trait for making live attenuated influenza vaccines. Temperature sensitivity generally occurs only after a series of substantial laboratory manipulations and selections. Interestingly, further investigation indicated the circulating strains in 1977-78 were often comprised of mixed temperature-sensitive and normal components, and that temperature sensitivity apparently disappeared from the post-1978 H1N1 lineage rapidly22. Escape of a mid-protocol population of H1N1 virus undergoing laboratory selection for temperature sensitive mutants would provide such a mixed population. In 1976-77 laboratory personnel in their late teens or early 20s would not have been exposed to pre-1957 H1N1 influenza viruses, and been susceptible to laboratory infections. The low severity of the 1977 pandemic might be in part due to the temperature sensitivity of the virus, a trait that limits virus replication in pulmonary tissues.
Example #3 Venezuelan Equine Encephalitis in 1995
Venezuelan Equine Encephalitis (VEE) is a viral disease transmitted by mosquitoes that intermittently erupts in regional or continental-scale outbreaks in the Western Hemisphere that involve equines (horses, donkeys and mules), termed epizootics, and often with concurrent epidemics among humans. The disease in equines creates high fever and severe neurological symptoms (colloquially termed “pesta loca” [crazy plague] in Spanish, or “blind staggers” in English) and a high, 19-83% fatality rate. In humans symptoms can vary from asymptomatic to a mild influenza-like febrile illness to a severe acute incapacitating febrile illness often with neurological symptoms (headache, depression, incoordination, mental clouding, epileptic seizures). Though its severity varies between outbreaks, VEE in humans may be fatal (up to ~5%) or, particularly in children, leave permanent neurological disability (epilepsy, paralysis, mental retardation) in 4 to 14% of clinical cases. In humans and equines miscarriages and stillbirths are increased.
Outbreaks typically occur in South America, though the 1969-71 continental-scale epizootic/epidemic reached from Central America through Mexico to Texas. VEE viruses are classified by their surface antigens, with the types causing large scale epizootics and epidemics being classed as type IAB and IC (termed epizootic strains), and types causing only sporadic human or equine disease in localized areas falling into types ID, IE, IF and II through VI (termed enzootic strains).
With modern genomic investigations available since the mid 1990s, programs of surveillance of mosquitoes and wildlife in regions at risk have discovered that in nature the enzootic VEE viruses are maintained by continuous transmission by mosquitoes in small mammals in the tropical and subtropical western hemisphere. The epizootic/epidemic type IAB and IC viruses appear suddenly without evidence of ongoing transmission during the long intervals between major outbreaks23. Moreover, genomic studies indicate that the epizootic types of VEE originate from the enzootic strains, specifically strain ID having given rise to the epizootic/epidemic types IAB and IC through a process of mutation24. VEE virus, like influenza virus shows rapid spontaneous changes in its genome, so that one can determine not only the genetic relatedness but also quantify the chronological distance separating different viral strains.
This is where the elegance of modern viral genomics becomes an embarrassment to virologists. It is clear from the genomics that while the enzootic type ID VEE virus can indeed mutate into the epizootic/epidemic types IAB and IC, it has, in fact, only done this on three occasions: ID to IAB some time in the 1930s and ID to IC in 1963 and 1992. There had been significant outbreaks of VEE every few years from the 1930s to the 1970s, however, and analysis showed that the numerous type IAB outbreaks were essentially matches to the original 1938 IAB VEE isolation that had been used in veterinary vaccines since the late 1930s. The veterinary vaccines had used inactivated (i.e. “killed”) whole virulent viruses. VEE is notoriously hard to inactivate in the lab, and laboratory infections were common. It was clear that many batches of the veterinary VEE vaccines had not been completely inactivated, in which residual infective virus remained.
From 1938 to 1972, the VEE vaccine was causing most of the very outbreaks that it was called upon to control, a vicious cycle indeed, and another example of “self-fulfilling prophecy” outbreaks.
The recognition that inadequately inactivated vaccines caused most VEE outbreaks caused a change in the veterinary vaccine seed virus to an attenuated strain, and VEE outbreaks apparently ceased for 20 years, from 1973 to 1992 25. Then, in 1992 a VEE outbreak in Venezuela occurred which proved to be a IC virus that was shown by genomic studies to have spontaneously arisen from enzootic type ID viruses circulating in the area where it arose, much like what had also occurred in the same area of Venezuela in 1962-64, when ID had mutated to a IC and caused an outbreak. The two IC VEE viruses, from 1962-64 and 1992, were distinct from each other, and arose from different genetic lines of ID viruses. The mystery of how epizootic VEE viruses arise naturally was apparently solved.
However, in 1995 a major VEE epizootic and epidemic hit Venezuela and Colombia, with a type IC virus also the cause. There were at least 10,000 human VEE cases with 11 deaths in Venezuela26 and an estimated 75,000 human cases in Colombia, with 3,000 neurological complications and 300 deaths27. Household attack rates ran 13-57% and VEE virus was isolated from 10 stillborn or miscarried human fetuses28.
Full genomic studies identified the 1995 virus as identical to an 1963 isolate with no sign that this virus had been circulating and the acquiring small genetic mutations indicative of replicating in hosts for 28 years. It was another case of “frozen evolution.” But it could not be another case of an outbreak caused by a defective inactivated VEE vaccine, because the 1963 type IC VEE virus had never been used to make a vaccine. Possible trans-ovarian transmission in mosquito vectors had been explored previously with negative results29. Suspicion fell on an inadvertent release from a virology lab, either by an unrecognized infection of a lab worker or visitor, or escape of an infected laboratory animal or mosquito. VEE is easily transmitted by the aerosol route during laboratory manipulations, and laboratory infections with VEE are common in unvaccinated persons. In this outbreak there was considerable circumstantial evidence for such a laboratory escape. The 1963 type IC VEE virus was used in an “inactivated” form as a reagent for testing purposes, and this reagent preparation was tested and was found to contain live virus. This reagent was used in the virology laboratory in Venezuela located where the 1995 outbreak first appeared, which was in an area without ongoing circulation of type ID enzootic viruses related to the 1963/1995 IC virus, and an area removed from where de novo IC VEE outbreaks had previously originated. Moreover, a report from this lab of an IC virus isolated from a surveillance mosquito pool in 1983 proved to be identical to the IC antigen strain, indicating a previous laboratory contamination event. The major scientific group working on VEE published a paper in 2001 stating the outbreak most likely was a laboratory escape, though this could not be proven30.
The situation becomes less clear-cut, because in 2005 the same group reported small outbreaks from 2000 and 2003 with multiple isolations of IC virus from equids in Venezuela, this time one identical to the 1995 virus31. Yet another example of “frozen evolution” but during a period when the 1963/1995 IC virus was no longer used widely as a reagent preparation, and it originated in an area with ongoing enzootic transmission of VEE viruses. The VEE working group backed off its earlier conclusion that the 1995 outbreak was likely laboratory mediated, but was unable to propose a natural process for the genomic stasis they reported.
The VEE working group clearly has great expertise, and one must respect their judgment that the 2000 isolations are valid and laboratory circumstances are significantly different than in 1995, so that natural genomic stasis may indeed exist for VEE and is worthy of further investigation. Several proposed mechanisms for genomic stasis for VEE have been proposed and investigated. VEE circulates in a complex ecological pattern, with enzootic transmission involving a variety of mosquito and mammalian hosts, so various theories allowing genomic stasis have been proposed, such as latency in an arthropod line or mammalian host. These have been investigated with multi-year surveys in enzootic and post-epizootic areas, and no definite evidence of persistent epizootic strains have been found in arthropods or small mammals. In addition to the negative surveys, the short lifespans of small mammals and of the potential arthropod hosts preclude viral latency from explaining the 5 or 28 year hiatuses in the appearances of the “frozen” IC epizootic viruses, and no evidence has been found for latency in the longer-lived human and equine hosts. Transovarian “vertical” propagation of viruses in arthropods between mother and progeny has been described with some pathogens in arthropods, and this has been investigated experimentally with VEE in VEE vectors, without positive results32.
It is clear that laboratory strains of VEE virus have a decades-long established habit of re-appearing showing “frozen evolution,” and causing “self-fulfilling prophecy” epidemics. It is clear that escape of laboratory strains of this virus through faulty vaccines has occurred multiple times in the past. Strong circumstantial evidence exists for an inadvertent escape in 1995, and a re-emergence in 2000 is without explanation.
Example 4: SARS laboratory escapes outbreaks after the SARS epidemic
The SARS outbreak of 2002-2003 eventually spread to 29 countries, causing over 8,000 infections and at least 774 deaths. Because many cases were in hospital workers (1707, amounting to 21%), it had the potential to shut down health care services where it struck33. By imposing strict (sometimes draconian) quarantines on exposed persons and isolation of patients, and even more because of good fortune and dedicated (indeed, heroic) medical personnel, it was contained and extinguished by July 2003. Quarantines, closure of factories and travel restrictions caused economic losses estimated at $40 billion worldwide, with an estimated 2.6% GDP loss in China, 1.05% GDP loss in Hong Kong, and 0.15% GDP loss to Canada34.
SARS is particularly dangerous to handle in the laboratory because there is no vaccine, so all laboratory workers are susceptible. It can be transmitted through aerosol/droplet mechanisms: the very large (321 cases) Amoy Gardens outbreak in Hong Kong was traced to infectious aerosols created by turbulent flushing water flow in the sewer lines: this turbulent flow generated aerosols that were sucked back up into numerous adjacent apartments through dry floor drains by negative pressure generated by bathroom exhaust fans! (Abraham 2005).
Moreover, about 5% of SARS patients are “super-spreaders” who pass the infection to many (over 8) secondary cases35. One case (ZZ) spread SARS to directly to 28 persons during one 18-hour hospitalization, before transfer to another hospital, where he infected 93 additional hospital personnel. At a third hospital he infected 23 staff and 19 patients, and at a fourth 20 hospital staff (Abraham 2005). Another super-spreader in Beijing infected at least 59 secondary cases. A super-spreader originally infected by ZZ in China visited Hong Kong but fell ill and remained in his hotel room, but managed to spread SARS to 10 secondary cases whose only associations were using a common elevator or hallway. These Hong Kong hotel exposures were international tourists, however, and were responsible for spreading SARS to Canada, Ireland, the US, Singapore, and Vietnam36. A 72-year old was already ill when he boarded flight CA112 from Hong Kong to Beijing on March 15, after having visited a niece ill with SARS in a Hong Kong hospital. Besides introducing another transmission chain in Beijing, on the two-hour flight he infected 20 other passengers and 2 flight attendants, who spread the disease to Mongolia, Singapore, Taiwan, and re-introduced new infection chains back into Hong Kong37 (Abraham 2005).
The existence of SARS “super-spreaders” makes even a single laboratory infection into a potential pandemic.
SARS has not naturally recurred, but there have been six separate “escapes” from virology labs studying it: one each in Singapore and Taiwan, and in four distinct events at the same laboratory in Beijing.
The first escape was in Singapore in August 2003, in a 27-year-old virology graduate student at the National University of Singapore. He had not worked directly with SARS, but SARS was present in the virology laboratory where he worked with West Nile Virus (WNV). Investigation showed that his preparation of WNV was contaminated with SARS virus, and that this was the likely origin of his infection. After falling ill on Aug 26, he sought outpatient medical care in several venues, and was admitted to the hospital only on September 3. Fortunately he recovered and there were no secondary cases. Investigation revealed multiple shortcomings in infrastructure, training and observed procedures at the laboratory, and remedial actions were ordered38.
The second escape was in Taiwan in December 2003, when a SARS research scientist fell ill on a return airflight after attending a medical meeting in Singapore Dec 7-10. Although he felt his illness was SARS, he remained at home for 5 days, unwilling to seek medical care because he dreaded bringing disgrace to himself and his institution. He was only persuaded to enter the hospital when his father threatened to commit suicide39. Preliminary investigation implicated a laboratory exposure due to an attempt to decontaminate a bag of leaking biological waste, perhaps without proper protection and against protocol the day before he left for Singapore40. His 74 contacts in Singapore were put under quarantine for ten days, but again, fortunately none developed SARS. An expert committee from WHO investigated the laboratory and its procedures, and recommended improvements41.
This second outbreak further shook the virology communities in Asia, where many labs held and worked on SARS samples. On December 18, 2003 WHO released a new protocol for handling SARS specimens in the post-outbreak period, with special emphasis on reducing risk of and performing surveillance to detect laboratory infections42. Although this protocol was clearly created after the first (Singapore) escape, WHO chose to parse its words to avoid offending members. Perhaps distinguishing between a primary laboratory infection and secondary spread into a community “outbreak,” it chose to treat the risk as hypothetical, stating in the introduction:
The hypothetical outbreak was not long in coming.
On April 22, 2004 China reported a suspected case of SARS in a 20-year-old nurse who fell ill April 5 in Beijing. The next day it reported she had nursed a 26-year-old female laboratory researcher who had fallen ill on March 25. Still ill, the researcher had traveled by train to her home in Anhui province where she was nursed by her mother, a physician, who fell ill on April 8 and died April 19. The researcher had worked at the Chinese National Institute of Virology (NIV) in Beijing, which is part of China’s Center for Disease Control (CDC), and which was a major center of SARS research. The investigation at NIV also uncovered an unrelated laboratory infection in a 31-year old male laboratory researcher at the NIV who fell ill on 17 April [43]. The entire NIV institute was closed and all of its 200 employees placed in quarantine in a hotel. Subsequent investigation confirmed these first three cases as SARS, and eventually identified a total of nine cases, in three generations, including health care workers and their family contacts44. Neither of the two primary patients had worked with live SARS virus, and WHO investigators had “serious concerns” regarding biosafety procedures at the NIV45.
Several Chinese and international groups investigated the outbreak at the NIV, and identified in retrospect two additional SARS laboratory infections at the NIV that had previously gone unrecognized and had begun in February 2004 [46]. A joint China CDC and WHO investigation found many shortcomings in biosecurity at the NIV, and traced the specific cause of the outbreak to an inadequately inactivated preparation of SARS virus that was used in general (not biosecure) laboratory areas in the NIV, including the one in which the two primary cases worked. It had not been tested to confirm its safety after inactivation, as it should have been. The WHO also found more general shortcomings in the handling of live SARS virus and a lack of surveillance of laboratory personnel for laboratory infections.
Li Liming, director of the China CDC and his deputy directory, the director of the NIV and his deputy director, and the director of the division where the two index cases worked were removed from their positions and found guilty of negligence in overseeing safety at the institution47. The Chinese government also decided to move the China CDC campus from its position in a residential neighborhood to an area “more remote from downtown,” and to allocate funds for more advanced laboratory equipment and infrastructure48.
Interestingly, the virology community is still reticent to discuss laboratory escapes: despite the considerable alarm these escapes created in the public health community and the participation of US CDC personnel in their investigation, they go unmentioned in the “10 years after” historical review of SARS by the CDC.49
Example 5: Foot and Mouth Disease (FMD) from Pirbright 2007
Foot and Mouth Disease (FMD) is a veterinary disease that affects primarily cloven-hoofed domestic animals (pigs, sheep and cattle). It has been eradicated in North America and most of Europe. It is highly transmissible, capable of spreading through direct contact and even through some prepared meats (sausages, airline food), on boots of farm workers (or tourists’ shoes: that’s why there’s that question “have you visited a farm” on the re-entry customs checklist coming into the USA), and even by aerosol spread.
FMD only occasionally causes a mild disease in humans, though exposed humans can carry the virus for up to three days, potentially an important method of spread. FMD causes a more serious disease in animals. Often fatal in young animals, survivors are stunted and lose their economic value. Adult animals die less often, but fail to gain weight or drop in milk production, and can become carriers. Most importantly, strict international quarantine regulations mean that an outbreak will cause all livestock and meat from that country to be banned from international trade. Various methods of outbreak control exist, but all are draconian, requiring massive culling (killing) of “in contact” but otherwise healthy animals surrounding index cases. Restrictions on all animal movement and often all commerce through infected areas are imposed, resulting in secondary economic losses from loss of tourism and general economic activity.
For instance, in the UK in 2001 a FMD outbreak ran from February to October 2001, with travel and export restrictions lasting into 2002. To control the outbreak, all susceptible livestock within 3 kms of an active case were culled. At its peak, 80,000-93,000 animals a week were killed and burned on farms, a total about 10 million sheep and cattle. Its direct cost was about $6.9 billion with overall costs to the British economy estimated at $16 billion.
On August 3, 2007 an outbreak of FMD was reported on a farm in the UK, initially with at least 38 cases in cattle identified. Quarantine measures were introduced and an investigation begun, with culling of surrounding livestock. Most countries banned UK livestock and meat exports. The virus was quickly identified as a strain that had caused a 1967 outbreak in the UK, but was not currently circulating in animals anywhere. Another case of “frozen evolution.” However, this outbreak was 2.8 miles (4.6 kms) south of Pirbright, where the only two facilities in the UK that were authorized to hold FMD virus were located. One was the UK Institute for Animal Health (IAH), the other Merial, a commercial veterinary FMD vaccine manufacturer. They both used the 1967 FMD strain, the Merial facility in large amounts (10,000 l) for vaccine manufacture. Operations were suspended at Merial on August 4 and its license to operate withdrawn. A second FMD outbreak quickly appeared near the first, and animal movement with the UK restricted and quarantine zones encompassing both the Pirbright campus and two affected farms were put in place on Aug 750. An initial investigation also published August 7 found no evidence for aerosol or surface water transmission of FMD virus from Pirbright, was investigating other wastewater issues, and suggested human carriage might have occurred51.
Investigation eventually showed that a waste-water line carrying partially treated waste water from the Merial vaccine plant to the final waste treatment plant run by IAH had gone without routine inspection or maintenance, was damaged, leaking, and had an unsealed manhole opening to the surface, so was capable of contaminating ground and surface water. It became clear that Merial and the IAH each considered the other responsible for such inspection and maintenance, and it had gone undone. The non-secure wastewater line ran through a construction area that recent heavy rains had turned into deep mud, and construction vehicles traversed it and exited the Pirbright campus without inspection or monitoring. These trucks sometimes used the road that passed by the first affected farm. It was concluded that contaminated mud from the defective wastewater line at Pirbright had been carried on tyres or underbody of construction vehicles and caused the first outbreak52.
For a brief time the outbreak was thought to have ended, and restrictions in the Pirbright area were lifted September 8, 2007. The UK applied to the EU to lift most restrictions on animal exports from the UK to EU on September 11, 2007.
However, on September 12, 2007 FMD was again reported, this time 30 miles north of Pirbright, again with the same 1967 strain of FMD. From September 18-30 multiple additional outbreaks of FMD appeared in the same area. A national embargo on all animal shipment was imposed, and new surveillance zones expanded rapidly until, overlapping they encompassed a portion of Heathrow Airport and were cut across by the major M4, M3 and M25 motorways. Rapid (real-time) genomic analysis had been ongoing during this outbreak, and indicated a single escape of FMD from Pirbright, which first spread between the two farms of the August outbreak, then went unnoticed at third farm before it blossomed again in mid September. Follow-up investigations identified the intermediate farm53.
The 2007 UK FMD outbreak identified 278 infected animals, and required 1578 animals to be culled54. It disrupted UK agricultural production and exports, and cost an estimated 200 million pounds. The ban of meat exports was particularly damaging as UK beef had only just exited a 10-year embargo by the EU because of BSE (Mad Cow Disease) in May of 2006.
FMD is such an easily transmitted virus with such potential to cause massive economic damage it would appear that manipulating it in a virology laboratory in a FMD free area is manifestly fraught with hazard. Particularly when it might escape by an “invisible” breach in biosafety as it did at Pirbright, and where it might lurk undetected despite heavy surveillance as it did between the two outbreaks.
In the US, previous law had banned it on the continental US, so FMD virus is currently only held in the USDA Plum Island facility off of Long Island (in a facility originally built in the 1950s for anti-animal BW [biological weapons] work). Currently a replacement facility under the Department of Homeland Security (DHS), the National Bio-and Agro-Defense Facility (NBAF) is under construction in Manhattan, KS. The move of FMD research to the agricultural heartland of the US was opposed by many groups, including the GAO, but DHS decided on the KS location and construction is ongoing. So much for learning from other’s experience.
Conclusions
There are some common themes in these narratives of escaped pathogens. Undetected flaws in the functioning of what was considered at the time to be an adequate standard of technical biocontainment is one theme, as demonstrated in the UK smallpox and FMD cases. Transfer to and handling of inadequately inactivated preparations of dangerous pathogens in areas of the laboratory with reduced biosecurity levels (allowable if the preparation is actually inactivated) is another theme, demonstrated in the SARS and VEE escapes. Poor training of personnel and slack oversight of laboratory procedures negated policy efforts by national and international bodies to achieve biosecurity in the SARS and UK smallpox escapes. The recent appearance of a cohort of immunologically naïve people in the general population, which previously had been uniformly immune was a factor in the UK smallpox and the 1977 H1N1 escapes; in this regard it should be remembered that there is no immunity at all in the general population to most potentially pandemic pathogens currently under discussion, such as Avian influenza and SARS.
It is hardly reassuring that despite stepwise technical improvements in containment facilities and increased policy demands for biosecurity procedures in the handling of dangerous pathogens, that escapes of these pathogens regularly occur and cause outbreaks in the general environment. Looking at the problem pragmatically, question is not if such escapes will happen in the future, but rather what the pathogen may be and how such an escape will be contained, if indeed it can be contained at all.
Advances in genetic manipulation now allow the augmentation of virulence and transmissibility in dangerous pathogens, and such experiments have been funded and performed, notably in the H5N1 avian influenza virus. The advisability of performing such experiments at all, and particularly in laboratories placed at universities in heavily populated urban areas, where laboratory personnel who are potentially exposed are in daily contact with a multitude of susceptible and unaware citizens is clearly in question.
If such manipulations should be allowed at all, it would seem prudent to conduct them in isolated laboratories where personnel are sequestered from the general public and must undergo a period of “exit quarantine” before re-entering civilian life55. Such isolated “detached duty”, while inconvenient for the lifestyle of virologists, is hardly foreign to them, since many experience prolonged periods of inconvenient and dangerous field work in the collection of viruses in the field, and certainly many other natural scientists do prolonged and isolated field work as well. The “inconvenience” barrier that requiring such isolation may present to principal investigators and other personnel may act as a natural screening factor to insure that dangerous manipulations to dangerous pathogens are only undertaken when genuinely indicated.
_______________
REFERENCES
1 Marc Lipsitch and Barry R. Bloom 2012. Rethinking Biosafety in Research on Potential Pandemic Pathogens . mBio 3(5): . doi:10.1128/mBio.00360-12.
2 Klotz, LC. The Human Fatality and Economic Burden of a Man-made Influenza Pandemic: A Risk Assessment. Unpublished Jan 2014. May be accessed at http://armscontrolcenter.org/The_Human_ ... 1-5-14.pdf or http://bio-security.org/wp-content/uplo ... 1-5-14.pdf
3 Merler S, Ajelli M, Fumanelli L, and Vespignani A. Containing the accidental laboratory escape of potential pandemic influenza viruses. BMC Medicine 2013, 11:252 doi:10.1186/1741-7015-11-252 http://www.biomedcentral.com/1741-7015/11/252
4 Scientists call for urgent talks on mutant-flu research in Europe. By Heidi Ledford. Nature 20 December 2013 http://www.nature.com/polopoly_fs/7.145 ... letter.pdf
5 Klotz LC, Sylvester EJ. The unacceptable risks of a man-made pandemic. Bulletin of the Atomic Scientists 7 Aug 2012.
6 Henkel R, Miller T, and Weyant RS. Monitoring Select Agent Theft, Loss and Release Reports in the United States—2004-2010. Applied Biosafety 17 (4) 2012 http://www.absa.org/abj/abj/121704FAHenkel.pdf Escaped Viruses-final 2-17-14
7 Baillie GJ, et al. Evolutionary dynamics of local Pandemic H1N1/2009 Influenza virus lineages revealed by whole-genome analysis. J Virol 86(1):11-18 Jan 2012.
8 Shooter RA. Report of the Investigation into the Cause of the 1978 Birmingham Smallpox Occurrence. HMSO 1980. Available: http://www.official-documents.gov.uk/do ... 8/0668.pdf
9 Fenner F. Henderson D. A. Et al. Smallpox and its Eradication. Published by World Health Organization, Geneva, 1988. ISBN 10: 9241561106 / ISBN 13: 9789241561105 available online: http://apps.who.int/iris/handle/10665/39485.
10 Kung HC, et al. Influenza in China in 1978: recurrence of influenza virus A subtype H1N1. Bull WHO 56(6):913-918 (1978).
11 Beveridge WIB. Where did red flu come from? New Sci. 1978 Mar 23;77/1095:790–1.
12 Nakajima K, Desselberger U, Palese P. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature. 1978;274:334–339.
13 Scholtissek C, von Hoyningen V, Rott R. Genetic relatedness between the new 1977 epidemic strains (H1N1) of influenza and human influenza strains isolated between 1947 and 1957 (H1N1). Virology. 1978;89:613–617
14 Chakraverty P. The return of the historic influenza A H1N1 virus and its impact on the population of the United Kingdom. J Hyg, Camb (1982) 89: 89-100
15 Shortridge KF. Et al. Reappearance of H1N1 influenza virus in man: evidence for the persistence of the virus in domestic chickens. Bull WHO 57(3):475-477 (1979)
16 Zhang G, Shoham D, Gilichinsky D, Davydov S, Castello JD, Rogers SO. Evidence of influenza a virus RNA in siberian lake ice. J Virol. 2006 Dec;80(24):12229-35. Epub 2006 Oct 11. Erratum in: J Virol. 2007 Mar;81(5):2538.
17 Gang Zhang, Dany Shoham, David Gilichinsky, Sergei Davydov, John D. Castello, and Scott O. Rogers. Correction: Evidence of influenza a virus RNA in Siberian lake ice. J Virol. 2007 March; 81(5): 2538. doi: 10.1128/JVI.02773-06 PMCID: PMC1865937
18 Worobey M.J. Phylogenetic evidence against evolutionary stasis and natural abiotic reservoirs of influenza A virus Virol. 2008 Apr;82(7):3769-74. doi: 10.1128/JVI.02207-07. Epub 2008 Jan 30.
19 Zimmer SM, Burke DS. Historical perspective–Emergence of influenza A (H1N1) viruses. N Engl J Med. 2009;361:279–285. [PubMed]
20 Nelson MI, Viboud C, Simonsen L, Bennett RT, Griesemer SB, St George K, Taylor J, Spiro DJ, Sengamalay NA, Ghedin E, Taubenberger JK, Holmes EC. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 2008 Feb 29;4(2):e1000012. doi: 10.1371/journal.ppat.1000012.
21 Wertheim JO. The re-emergence of H1N1 influenza virus in 1977: a cautionary tale for estimating divergence times using biologically unrealistic sampling dates. PLoS One. 2010 Jun 17;5(6):e11184. doi: 10.1371/journal.pone.0011184.
22 Oxford JS. Et al. Naturally occurring temperature-sensitive Influenza A viruses of the H1N1 and H3N2 subtypes. J Gen Virol (1980). 48, 383-389.
23 Scherer WF, et al. Vector incompetency: Its implication in the disappearance of epizootic Venezuelan equine encephalomyelitis virus form Middle America. J Med Etomol 23(1):23-29 1986 Jan 24.
24 Powers AM, et al. Repeated emergence of epidemic/epizootic Venezuelan equine encephalitis fro a single genotype of enzootic subtype ID virus. J Virol 71(9):6697-6705. Sept 1997.
25 Weaver, SC, et al. Genetic evidence for the origins of Venezuelan equine encephalitis virus subtype IAB outbreaks. Am J Trop Med Hyg 60(30): 441-448. 1999.
26 Braulty AC, et al. Potential sources of the 1995 Venezuelan equine encephalitis subtype IC epidemic. J Virol 75(13): 5823-5832 July 2001.
27 Rivas F, et al. Epidemic Venezuelan equine encephalitis in La Guajira, Colombia, 1995. J Infect Dis 1997;175:828-832.
28 Weaver SC, et al. Re-emergence of epidemic Venezuelan equine encephalomyelitis in South America. Lancet 1996 Aug 17 348(9025):346-340.
29 Scherer WF, et al. Vector incompetency: Its implication in the disappearance of epizootic Venezuelan equine encephalomyelitis virus from Middle America. J Med Etomol 23(1):23-29 1986 Jan 24.
30 Braulty AC, et al. Potential sources of the 1995 Venezuelan equine encephalitis subtype IC epidemic. J Virol 75(13): 5823-5832 July 2001.
31 Navarro J-C, et al. Postepizootic persistence of Venezuelan equine encephalitis virus, Venezuela. Emerg Infect Dis 11(12):1907-1915 Dec 2005.
32 Scherer WF, et al. Vector incompetency: Its implication in the disappearance of epizootic Venezuelan equine encephalomyelitis virus from Middle America. J Med Etomol 23(1):23-29 1986 Jan 24.
33 Abraham, T. Twenty-first Century Plague: The Story of SARS. Baltimore MD: Johns Hopkins Univ Press 2005. Is a short, useful summary of the SARS epidemic.
34 International Dimensions of Ethics Education in Science and Engineering: Case Study Series. IDEESE: Cases – Reporting Incidence of SARS: Appendix A. available at: http://www.umass.edu/sts/ethics/sars.html.
35 Zhuaang Shen, et al. Superspreading SARS events, Beijing, 2003 Emerg Infect Dis 2004 10(2):256-260. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3322930/#R1
36 MMWR. Update: Outbreak of Severe Acute Respiratory Syndrome – Worldwide, 2003. MMWR March 28, 2003 / 52(12);241-248. http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5212a1.htm
37 WHO. World Health Report 2007 Ch 3: (illustration): Probable SARS transmission on flight CA112 in March 2003. http://www.who.int/whr/2007/media_centr ... g01_en.pdf
38 Singapore Ministry of Health. Biosafety and SARS Incident in Singapore September 2003. Websource: http://www.biosafety.be/CU/PDF/Report_S ... gapore.pdf
39 WHO Global Alert and Response: Severe Acute Respiratory Syndrome (SARS) in Taiwan, China. 17 Dec 2003. http://www.who.int/csr/don/2003_12_17/en/
40 Reuters: Taipei, Dec 20,2003. Taiwan says scientist likely got SARS in lab slip. Accessed at: http://www.royalsociety.org.nz/2003/12/ ... -taiwan-2/
41 Wu, W C et al. Development of Laboratory Biosafety Management: The Taiwan Experience. Applied Biosafety 12(1):18-25 2007. Accessed at: http://www.absa.org/abj/abj/071201wu.pdf
42 WHO. WHO post-outbreak biosafety guidelines for handling of SARS-CoV specimens and cultures. 18 Dec 2003. Accessible at: http://www.who.int/csr/sars/biosafety2003_12_18/en/
43 WHO Global Alert and Response (GAR). China reports additional SARS cases – update 23 April 2004. http://www.who.int/csr/don/2004_04_23/en/index.html
44 WHO Global Alert and Response (GAR). China confirms SARS in infection in another previously reported case; summary of cases to date – update 5. 30 April 2004. http://www.who.int/csr/don/2004_04_30/en/index.html
45 WHO Global Alert and Response (GAR). China’s latest SARS outbreak has been contained, but biosafety concerns remain – update 7. 18May2004. http://www.who.int/csr/don/2004_05_18a/en/index.html
46 Zhao Xiaojian, Zhu Xiaochao. China CDC Blamed for SARS escape. Caijing.com.cn Daily Update May 20, 2004. http://english.caijing.com.cn/2004-05-20/100013918.html
47 SCiDevNet website. Top Chinese Scientists ‘punished’ over SARS outbreak. July 8, 2004. http://www.scidev.net/global/health/new ... utbre.html
48 Zhang Feng (China Daily). Officials punished for SARS virus leak. Chinadaily.com.cn July 7, 2004. http://www.chinadaily.com.cn/english/do ... 344755.htm
49 CDC website. Remembering SARS: A deadly puzzle and the efforts to solve it. Online source: http://www.cdc.gov/about/history/sars/feature.htm
50 DEFRA. Declaration of a Protection Zone and Surveillance Zone 7Aug2007. http://webarchive.nationalarchives.gov. ... rz0807.pdf
51 HSE. Initial report on potential breaches of biosecurity at the Pirbright site 2007. Web source: http://webarchive.nationalarchives.gov. ... 0807b.htm/
52 Logan, P. Final report on potential breaches of biosecurity at the Pirbright site 2007. Health and Safety Executive September 2007. http://www.hse.gov.uk/news/2007/finalreport.pdf
53 Cottam EM, et al. Transmission pathways of Food-and-Mouth Disease Virus in the United Kingdom in 2007. PLosPathogens April 2008 4(4) e1000050. Web: http://www.plospathogens.org/article/in ... at.1000050
54 Juleff, N. The 2007 UK FMD outbreak: field investigation perspective. Institute for Animal Health (UK). PowerPoint presentation. Available online: http://www.fao.org/ag/againfo/commissio ... ctives.pdf
55 Klotz, LC and Sylvester, E. The unacceptable risks of a man-made pandemic. Bulletin of the Atomic Scientists. 7 August 2012. http://thebulletin.org/unacceptable-ris ... e-pandemic
by Martin Furmanski MD
Scientist’s Working Group on Chemical and Biologic Weapons
Center for Arms Control and Nonproliferation
February 17, 2014
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
YOU ARE REQUIRED TO READ THE COPYRIGHT NOTICE AT THIS LINK BEFORE YOU READ THE FOLLOWING WORK, THAT IS AVAILABLE SOLELY FOR PRIVATE STUDY, SCHOLARSHIP OR RESEARCH PURSUANT TO 17 U.S.C. SECTION 107 AND 108. IN THE EVENT THAT THE LIBRARY DETERMINES THAT UNLAWFUL COPYING OF THIS WORK HAS OCCURRED, THE LIBRARY HAS THE RIGHT TO BLOCK THE I.P. ADDRESS AT WHICH THE UNLAWFUL COPYING APPEARED TO HAVE OCCURRED. THANK YOU FOR RESPECTING THE RIGHTS OF COPYRIGHT OWNERS.
Introduction
The danger to world or regional public health from the escape from microbiology laboratories of pathogens capable of causing pandemics, or Potentially Pandemic Pathogens (PPPs) has been the subject of considerable discussion1,2,3,4 including mathematical modeling of the probability and impact of such escapes5. The risk of such releases has generally been determined from estimates of laboratory infections that are often incomplete, except for the recent 2013 Centers for Disease Control (CDC) report6, which is a significant source of recent data on escapes from undetected and unreported laboratory-acquired infections (LAIs).
This paper presents an historical review of outbreaks of PPPs or similarly transmissible pathogens that occurred from presumably well-funded and supervised nationally supported laboratories. It should be emphasized that these examples are only the “tip of the iceberg” because they represent laboratory accidents that have actually caused illness outside of the laboratory in the general public environment. The list of laboratory workers who have contracted potentially contagious infections in microbiology labs but did not start community outbreaks is much, much longer. The examples here are not “near misses;” these escapes caused real-world outbreaks.
Methods of pathogen identification
Modern genetic analysis allows pathogens to be identified, and given a sufficient catalog of isolates of the same pathogen, it is possible to determine if two specimens are identical or very closely related. Because all pathogens that are circulating in the environment show genetic changes over time, one can date the time the pathogen circulated. For instance, for 20th century human and swine influenza viruses beginning in the 1930s, one can generally place a virus to a particular year. With modern rapid genomic analysis outbreaks can be traced with considerable accuracy: for instance the 2009 pandemic pH1N1 influenza outbreak has been analyzed with confidence limits of branchpoints in its first wave defined within days or weeks, and individual transmission chains can be identified7.
Example #1: British smallpox escapes, 1966, 1972, 1978
The WHO’s successful effort to eradicate natural transmission of smallpox in the 1970s highlighted the risk that virology laboratories posed as a source of epidemics. This was clearly demonstrated in the United Kingdom, where from 1963-1978 only 4 cases of smallpox (with no deaths) were reported from smallpox endemic areas, while during the same period at least 80 cases and 3 deaths were the result of three separate escapes of the smallpox virus from two different accredited smallpox laboratories.8 Much of the current policy and practice in biosafety and biocontainment of dangerous pathogens can be traced to the political and professional reaction to these outbreaks.
The UK became a sensitive test system for smallpox laboratory escapes because it ended compulsory smallpox vaccination in 1946. Public sentiment in the UK had always included significant resistance to and apathy towards vaccination, and so by the mid 1960s and through the 1970s a large proportion of children and young adults had never been vaccinated, and many older persons were never re-vaccinated after initial childhood or military vaccinations. Thus the protective herd immunity in the general public, which earlier rendered impotent any laboratory escapes, disappeared. At the same time, the considerable volume of travel and immigration from smallpox endemic areas of Africa and the Indian subcontinent meant that surveillance for imported smallpox cases was required. UK maintained several smallpox laboratories at medical schools for both research and to support clinical diagnosis.
The first laboratory outbreak to be recognized began in March 1972, in a 23 year old laboratory assistant at the London School of Hygiene and Tropical Medicine, who had observed harvesting of live smallpox virus from eggs. This had been done on an open bench, as was routine, the laboratory having no isolation cabinets at that time. Before she was placed in isolation, she infected two visitors to a patient in an adjacent bed, both of whom died. They in turn infected a nurse, who survived9.
The recognition of this laboratory escape resulted in several investigations, which led to the establishment of guidelines for laboratories handling smallpox and other dangerous pathogens. These recommendations included handling dangerous pathogens in biological safety cabinets only in certain dedicated rooms by specifically trained and designated personnel. Also guidelines were issued for isolation with dedicated gowns and gloves, and the establishment of proper ventilation facilities to maintain negative pressure in these rooms and cabinets. These recommendations are the direct precursors to the current Biosafety Laboratory (BSL) level protocols.
By 1977 the natural chain of smallpox transmission had been interrupted, and the WHO was in the process of reducing the number of laboratories holding smallpox virus. In August of 1978 a 40 year old medical photographer at Birmingham Medical School developed smallpox, and died. She infected her mother, who survived. She worked in a studio and darkroom that was immediately above the smallpox laboratory at Birmingham Medical School. Investigation revealed that although the long established laboratory had been inspected and approved to handle smallpox virus, it did not have sufficient facilities to meet the new biocontainment requirements, and was scheduled to be decommissioned at the end of 1978. Moreover, work on smallpox had accelerated substantially in order to complete existing projects before the closing, and work with smallpox was performed by laboratory personnel who did not receive appropriate training and supervision, and appropriate isolation practices were frequently violated. The most likely route of exposure of the Medical Photographer was by transport of infectious aerosols generated by a centrifuge through building ventilation ducts that were improperly sealed and allowed aerosols to be delivered to one of the Photographer’s working spaces. Laboratory notebooks and the photographer’s work logs indicated that the strain infecting the photographer was handled in the laboratory on the same days that the photographer worked in the potentially contaminated workspace, on dates consistent with the photographer’s calculated exposure date. Dr Henry Bedson, a world renowned smallpox investigator who was responsible for the Birmingham laboratory, committed suicide as a result of the outbreak (Shooter 1980).
The 1978 investigation re-examined a 1966 smallpox outbreak, which in retrospect was strikingly similar to the 1978 outbreak. The earliest case identified in 1966 was in a medical photographer who worked at Birmingham Medical School in the same facility as the 1978 case. This outbreak was caused by a low-virulence strain of smallpox (variola minor), and it caused at least 72 cases of smallpox from February to August 1966, spread through the midlands of Britain, and Wales. The vast majority of cases were in unvaccinated children or young adults. There were no deaths. Retrospective review again revealed variola minor had been manipulated in the smallpox laboratory at a time appropriate to cause the infection in the photographer working a floor above.
Example #2: The “re-emergence” of H1N1 human influenza in 1977.
Human influenza H1N1 viruses appeared with the 1918 pandemic, and persisted, slowing accumulating small changes in its genome (with a major change in 1947), until the H2N2 “Asian” flu appeared in 1957, causing a worldwide pandemic. H1N1 influenza virus then apparently became extinct, and was not isolated for 20 years. In 1969 the “Hong Kong” H3N2 virus replaced the H2N2 virus, and is still circulating.
In September 1977 an H1N1 influenza virus was isolated from human infections in the Far East region of the Soviet Union, and in early 1978 the Chinese reported they had isolated H1N1 virus in May of 1977 in northeast China adjacent to the Soviet outbreak1011. Using the early genetic tools available at the time, the 1977 H1N1 virus was found to be closely related to H1N1 human influenza viruses circulating in 1949-1950, but not to those circulating earlier or later12, 13.
The 1977 H1N1 flu virus rapidly spread worldwide, in a pandemic that was restricted largely to people under ~ 21 years of age. Older persons had been exposed to related H1N1 viruses prior to 1957, and carried substantial immunity. Mercifully, the “re-emergent” H1N1 virus was not very virulent. Although illness was widespread, affecting 20-70% of those under 20 years of age in school or military camp outbreaks in the first year14, deaths were few. Many asymptomatic infections were detected by serology (Kung 1978).
The appearance of this “time-traveling throwback” puzzled virologists, because no similar examples had previously been identified in influenza or other similar viruses. Initially escape of a virus kept in storage from c1950 from a virology lab was discussed, but such a laboratory accident was denied by Chinese and Soviet virologists (Kung 1978, Beveridge 1978). Western virologists quietly let the matter of a laboratory escape origin for the 1977 H1N1 virus drop from discussion, out of an abundance of scientific caution, and also out of an eagerness not to offend the Russian and Chinese scientists, whose early gestures of cooperation in worldwide influenza surveillance system were very important to foster, because such cooperation would allow tracking influenza globally.
Discussions of the origins of the 1977 H1N1 gave rise to hypotheses of natural “biological stasis” or viral latency in an undefined animal. Experimental investigations of possible transmission of human H1N1 viruses in avians were pursued, but with minimal success and no demonstration of persistent avian transmission15, nor were human viruses identified in avians in very extensive subsequent surveys. The ambiguous term “frozen evolution” was coined, allowing for the freezing to be biologically functional, metaphorical, refrigerative, or natural.
A 2006 paper16 claimed to have isolated H1N1 influenza virus RNA from ice and meltwater from Siberian lakes that were frequented by migratory birds. Since migratory birds naturally carry and shed a wide variety of influenza viruses, and since year to year variations in the amount of thawing of lake ice might allow influenza viruses shed from the migratory birds to remain physically frozen for a number of years, the paper stated this might be the mechanism for the re-emergence of the 1977 H1N1 flu. It emphasized the 1977 H1N1 link because the RNA sequences it reported isolating from the lakes were closely related to sequences of three H1N1 reference viruses that it characterized as being of avian origin that circulated in the late 1960s.
Problems soon arose with this paper, however. The authors issued a correction17 in 2007 indicating the H1N1 reference strains originally characterized as avian and from the 1960s were in fact of human origin, and dated from the 1930s. A paper highly critical of the 2006 Siberian Lake paper was published in 2008 18, presenting strong evidence that the reported isolation of influenza RNA from nature was the result of contamination in the laboratory by the standard reference strain of human H1N1 virus (isolated in 1933) that was used as a positive control in that laboratory.
Presently, with detailed sophisticated genomic analysis available, and with 32 years of circulation of the 1977 H1N1 virus available for study, no evidence of natural genomic stasis has been identified. It has become clear that its appearance in 1977 was almost certainly due to escape from a virology lab of a virus sample that had been frozen since c1950. Only since 2008 have virologists actually begun to make the suggestion of a probable laboratory release in scientific papers: “The reemergence in 1977 is unexplained and probably represents reintroduction to humans from a laboratory source19,” and “…little A/H1N1 evolution is evident over the twenty-year period of the virus’s global disappearance, supporting earlier suggestions that this subtype was most likely accidentally reintroduced into human circulation from a laboratory environment20.” It should be noted that this paper calculates the 1977 H1N1 virus had been circulating for 1 year before it was reported, so that geographic origin cannot be stated with certainty.
Only since 2009-2010 did major papers begin to state directly the 1977 emergence of H1N1 influenza was a laboratory related release: “The most famous case of a released laboratory strain is the re-emergent H1N1 influenza A virus which was first observed in China in May of 1977 and in Russia shortly thereafter21.” The paper made this statement in part because the continued “agnostic” approach to the 1977 re-emergence introduced unacceptable errors in calculating the genomic divergence dates for influenza virus strains.
Public awareness of the 1977 H1N1 pandemic and its likely laboratory origins has been virtually absent. Virologists and public health officials with the appropriate sophistication were quickly aware that a laboratory release was the most likely origin, but they were content not to publicize this, aware that such embarrassing allegations would likely end the then nascent cooperation of Russian and Chinese virologists, which was vital to worldwide influenza surveillance. An abundance of caution in making such suggestions was also in their own self-interest. The 1976 “swine flu” alarm and subsequent immunization program that proved to be unneeded caused 532 cases of Guillain-Barre syndrome and 32 deaths. It was widely considered a misadventure, and had severely damaged the public and political credibility of the virology and public health communities. An acknowledgement of a pandemic originating from their laboratories would have only worsened it. The most plausible reason for a Chinese or Russian laboratory to thaw out and begin growing a c1950 H1N1 virus in 1976-77 was as a response to the US 1976 “swine flu” program, which resulted in a program to immunize the entire US population against H1N1 influenza virus. It was clearly a rational response for other countries with virology capabilities to explore making their own H1N1 vaccines. Thawing available frozen stocks of virus was necessary, because H1N1 was no longer circulating. Modern commentators have begun to articulate this connection between the 1976 Swine flu immunization program and the 1977 H1N1 re-emergence:
“Perhaps an even more serious consequence [of the 1976 swine flu episode] was the accidental release of human-adapted influenza A (H1N1) virus from a research study, with subsequent resurrection and global spread of this previously extinct virus, leading to what could be regarded as a ‘self-fulfilling prophecy’ epidemic.” (Zimmer 2009)
The speculation that the 1977 release may have been related to H1N1 vaccine research is supported by the observation that in the initial outbreaks in China, nine of the ten viral isolates expressed “temperature sensitivity” (Kung 1978). Temperature sensitivity normally an uncommon trait, but one that was in the 1970s (and still is) a fundamental trait for making live attenuated influenza vaccines. Temperature sensitivity generally occurs only after a series of substantial laboratory manipulations and selections. Interestingly, further investigation indicated the circulating strains in 1977-78 were often comprised of mixed temperature-sensitive and normal components, and that temperature sensitivity apparently disappeared from the post-1978 H1N1 lineage rapidly22. Escape of a mid-protocol population of H1N1 virus undergoing laboratory selection for temperature sensitive mutants would provide such a mixed population. In 1976-77 laboratory personnel in their late teens or early 20s would not have been exposed to pre-1957 H1N1 influenza viruses, and been susceptible to laboratory infections. The low severity of the 1977 pandemic might be in part due to the temperature sensitivity of the virus, a trait that limits virus replication in pulmonary tissues.
Example #3 Venezuelan Equine Encephalitis in 1995
Venezuelan Equine Encephalitis (VEE) is a viral disease transmitted by mosquitoes that intermittently erupts in regional or continental-scale outbreaks in the Western Hemisphere that involve equines (horses, donkeys and mules), termed epizootics, and often with concurrent epidemics among humans. The disease in equines creates high fever and severe neurological symptoms (colloquially termed “pesta loca” [crazy plague] in Spanish, or “blind staggers” in English) and a high, 19-83% fatality rate. In humans symptoms can vary from asymptomatic to a mild influenza-like febrile illness to a severe acute incapacitating febrile illness often with neurological symptoms (headache, depression, incoordination, mental clouding, epileptic seizures). Though its severity varies between outbreaks, VEE in humans may be fatal (up to ~5%) or, particularly in children, leave permanent neurological disability (epilepsy, paralysis, mental retardation) in 4 to 14% of clinical cases. In humans and equines miscarriages and stillbirths are increased.
Outbreaks typically occur in South America, though the 1969-71 continental-scale epizootic/epidemic reached from Central America through Mexico to Texas. VEE viruses are classified by their surface antigens, with the types causing large scale epizootics and epidemics being classed as type IAB and IC (termed epizootic strains), and types causing only sporadic human or equine disease in localized areas falling into types ID, IE, IF and II through VI (termed enzootic strains).
With modern genomic investigations available since the mid 1990s, programs of surveillance of mosquitoes and wildlife in regions at risk have discovered that in nature the enzootic VEE viruses are maintained by continuous transmission by mosquitoes in small mammals in the tropical and subtropical western hemisphere. The epizootic/epidemic type IAB and IC viruses appear suddenly without evidence of ongoing transmission during the long intervals between major outbreaks23. Moreover, genomic studies indicate that the epizootic types of VEE originate from the enzootic strains, specifically strain ID having given rise to the epizootic/epidemic types IAB and IC through a process of mutation24. VEE virus, like influenza virus shows rapid spontaneous changes in its genome, so that one can determine not only the genetic relatedness but also quantify the chronological distance separating different viral strains.
This is where the elegance of modern viral genomics becomes an embarrassment to virologists. It is clear from the genomics that while the enzootic type ID VEE virus can indeed mutate into the epizootic/epidemic types IAB and IC, it has, in fact, only done this on three occasions: ID to IAB some time in the 1930s and ID to IC in 1963 and 1992. There had been significant outbreaks of VEE every few years from the 1930s to the 1970s, however, and analysis showed that the numerous type IAB outbreaks were essentially matches to the original 1938 IAB VEE isolation that had been used in veterinary vaccines since the late 1930s. The veterinary vaccines had used inactivated (i.e. “killed”) whole virulent viruses. VEE is notoriously hard to inactivate in the lab, and laboratory infections were common. It was clear that many batches of the veterinary VEE vaccines had not been completely inactivated, in which residual infective virus remained.
From 1938 to 1972, the VEE vaccine was causing most of the very outbreaks that it was called upon to control, a vicious cycle indeed, and another example of “self-fulfilling prophecy” outbreaks.
The recognition that inadequately inactivated vaccines caused most VEE outbreaks caused a change in the veterinary vaccine seed virus to an attenuated strain, and VEE outbreaks apparently ceased for 20 years, from 1973 to 1992 25. Then, in 1992 a VEE outbreak in Venezuela occurred which proved to be a IC virus that was shown by genomic studies to have spontaneously arisen from enzootic type ID viruses circulating in the area where it arose, much like what had also occurred in the same area of Venezuela in 1962-64, when ID had mutated to a IC and caused an outbreak. The two IC VEE viruses, from 1962-64 and 1992, were distinct from each other, and arose from different genetic lines of ID viruses. The mystery of how epizootic VEE viruses arise naturally was apparently solved.
However, in 1995 a major VEE epizootic and epidemic hit Venezuela and Colombia, with a type IC virus also the cause. There were at least 10,000 human VEE cases with 11 deaths in Venezuela26 and an estimated 75,000 human cases in Colombia, with 3,000 neurological complications and 300 deaths27. Household attack rates ran 13-57% and VEE virus was isolated from 10 stillborn or miscarried human fetuses28.
Full genomic studies identified the 1995 virus as identical to an 1963 isolate with no sign that this virus had been circulating and the acquiring small genetic mutations indicative of replicating in hosts for 28 years. It was another case of “frozen evolution.” But it could not be another case of an outbreak caused by a defective inactivated VEE vaccine, because the 1963 type IC VEE virus had never been used to make a vaccine. Possible trans-ovarian transmission in mosquito vectors had been explored previously with negative results29. Suspicion fell on an inadvertent release from a virology lab, either by an unrecognized infection of a lab worker or visitor, or escape of an infected laboratory animal or mosquito. VEE is easily transmitted by the aerosol route during laboratory manipulations, and laboratory infections with VEE are common in unvaccinated persons. In this outbreak there was considerable circumstantial evidence for such a laboratory escape. The 1963 type IC VEE virus was used in an “inactivated” form as a reagent for testing purposes, and this reagent preparation was tested and was found to contain live virus. This reagent was used in the virology laboratory in Venezuela located where the 1995 outbreak first appeared, which was in an area without ongoing circulation of type ID enzootic viruses related to the 1963/1995 IC virus, and an area removed from where de novo IC VEE outbreaks had previously originated. Moreover, a report from this lab of an IC virus isolated from a surveillance mosquito pool in 1983 proved to be identical to the IC antigen strain, indicating a previous laboratory contamination event. The major scientific group working on VEE published a paper in 2001 stating the outbreak most likely was a laboratory escape, though this could not be proven30.
The situation becomes less clear-cut, because in 2005 the same group reported small outbreaks from 2000 and 2003 with multiple isolations of IC virus from equids in Venezuela, this time one identical to the 1995 virus31. Yet another example of “frozen evolution” but during a period when the 1963/1995 IC virus was no longer used widely as a reagent preparation, and it originated in an area with ongoing enzootic transmission of VEE viruses. The VEE working group backed off its earlier conclusion that the 1995 outbreak was likely laboratory mediated, but was unable to propose a natural process for the genomic stasis they reported.
The VEE working group clearly has great expertise, and one must respect their judgment that the 2000 isolations are valid and laboratory circumstances are significantly different than in 1995, so that natural genomic stasis may indeed exist for VEE and is worthy of further investigation. Several proposed mechanisms for genomic stasis for VEE have been proposed and investigated. VEE circulates in a complex ecological pattern, with enzootic transmission involving a variety of mosquito and mammalian hosts, so various theories allowing genomic stasis have been proposed, such as latency in an arthropod line or mammalian host. These have been investigated with multi-year surveys in enzootic and post-epizootic areas, and no definite evidence of persistent epizootic strains have been found in arthropods or small mammals. In addition to the negative surveys, the short lifespans of small mammals and of the potential arthropod hosts preclude viral latency from explaining the 5 or 28 year hiatuses in the appearances of the “frozen” IC epizootic viruses, and no evidence has been found for latency in the longer-lived human and equine hosts. Transovarian “vertical” propagation of viruses in arthropods between mother and progeny has been described with some pathogens in arthropods, and this has been investigated experimentally with VEE in VEE vectors, without positive results32.
It is clear that laboratory strains of VEE virus have a decades-long established habit of re-appearing showing “frozen evolution,” and causing “self-fulfilling prophecy” epidemics. It is clear that escape of laboratory strains of this virus through faulty vaccines has occurred multiple times in the past. Strong circumstantial evidence exists for an inadvertent escape in 1995, and a re-emergence in 2000 is without explanation.
Example 4: SARS laboratory escapes outbreaks after the SARS epidemic
The SARS outbreak of 2002-2003 eventually spread to 29 countries, causing over 8,000 infections and at least 774 deaths. Because many cases were in hospital workers (1707, amounting to 21%), it had the potential to shut down health care services where it struck33. By imposing strict (sometimes draconian) quarantines on exposed persons and isolation of patients, and even more because of good fortune and dedicated (indeed, heroic) medical personnel, it was contained and extinguished by July 2003. Quarantines, closure of factories and travel restrictions caused economic losses estimated at $40 billion worldwide, with an estimated 2.6% GDP loss in China, 1.05% GDP loss in Hong Kong, and 0.15% GDP loss to Canada34.
SARS is particularly dangerous to handle in the laboratory because there is no vaccine, so all laboratory workers are susceptible. It can be transmitted through aerosol/droplet mechanisms: the very large (321 cases) Amoy Gardens outbreak in Hong Kong was traced to infectious aerosols created by turbulent flushing water flow in the sewer lines: this turbulent flow generated aerosols that were sucked back up into numerous adjacent apartments through dry floor drains by negative pressure generated by bathroom exhaust fans! (Abraham 2005).
Moreover, about 5% of SARS patients are “super-spreaders” who pass the infection to many (over 8) secondary cases35. One case (ZZ) spread SARS to directly to 28 persons during one 18-hour hospitalization, before transfer to another hospital, where he infected 93 additional hospital personnel. At a third hospital he infected 23 staff and 19 patients, and at a fourth 20 hospital staff (Abraham 2005). Another super-spreader in Beijing infected at least 59 secondary cases. A super-spreader originally infected by ZZ in China visited Hong Kong but fell ill and remained in his hotel room, but managed to spread SARS to 10 secondary cases whose only associations were using a common elevator or hallway. These Hong Kong hotel exposures were international tourists, however, and were responsible for spreading SARS to Canada, Ireland, the US, Singapore, and Vietnam36. A 72-year old was already ill when he boarded flight CA112 from Hong Kong to Beijing on March 15, after having visited a niece ill with SARS in a Hong Kong hospital. Besides introducing another transmission chain in Beijing, on the two-hour flight he infected 20 other passengers and 2 flight attendants, who spread the disease to Mongolia, Singapore, Taiwan, and re-introduced new infection chains back into Hong Kong37 (Abraham 2005).
The existence of SARS “super-spreaders” makes even a single laboratory infection into a potential pandemic.
SARS has not naturally recurred, but there have been six separate “escapes” from virology labs studying it: one each in Singapore and Taiwan, and in four distinct events at the same laboratory in Beijing.
The first escape was in Singapore in August 2003, in a 27-year-old virology graduate student at the National University of Singapore. He had not worked directly with SARS, but SARS was present in the virology laboratory where he worked with West Nile Virus (WNV). Investigation showed that his preparation of WNV was contaminated with SARS virus, and that this was the likely origin of his infection. After falling ill on Aug 26, he sought outpatient medical care in several venues, and was admitted to the hospital only on September 3. Fortunately he recovered and there were no secondary cases. Investigation revealed multiple shortcomings in infrastructure, training and observed procedures at the laboratory, and remedial actions were ordered38.
The second escape was in Taiwan in December 2003, when a SARS research scientist fell ill on a return airflight after attending a medical meeting in Singapore Dec 7-10. Although he felt his illness was SARS, he remained at home for 5 days, unwilling to seek medical care because he dreaded bringing disgrace to himself and his institution. He was only persuaded to enter the hospital when his father threatened to commit suicide39. Preliminary investigation implicated a laboratory exposure due to an attempt to decontaminate a bag of leaking biological waste, perhaps without proper protection and against protocol the day before he left for Singapore40. His 74 contacts in Singapore were put under quarantine for ten days, but again, fortunately none developed SARS. An expert committee from WHO investigated the laboratory and its procedures, and recommended improvements41.
This second outbreak further shook the virology communities in Asia, where many labs held and worked on SARS samples. On December 18, 2003 WHO released a new protocol for handling SARS specimens in the post-outbreak period, with special emphasis on reducing risk of and performing surveillance to detect laboratory infections42. Although this protocol was clearly created after the first (Singapore) escape, WHO chose to parse its words to avoid offending members. Perhaps distinguishing between a primary laboratory infection and secondary spread into a community “outbreak,” it chose to treat the risk as hypothetical, stating in the introduction:
“The possibility that a SARS outbreak could occur following a laboratory accident is a risk of considerable importance, given the relatively large number of laboratories currently conducting research using the SARS-CoV or retaining specimens from SARS patients. These laboratories currently represent the greatest threat for renewed SARS-CoV transmission through accidental exposure associated with breaches in laboratory biosafety.”
The hypothetical outbreak was not long in coming.
On April 22, 2004 China reported a suspected case of SARS in a 20-year-old nurse who fell ill April 5 in Beijing. The next day it reported she had nursed a 26-year-old female laboratory researcher who had fallen ill on March 25. Still ill, the researcher had traveled by train to her home in Anhui province where she was nursed by her mother, a physician, who fell ill on April 8 and died April 19. The researcher had worked at the Chinese National Institute of Virology (NIV) in Beijing, which is part of China’s Center for Disease Control (CDC), and which was a major center of SARS research. The investigation at NIV also uncovered an unrelated laboratory infection in a 31-year old male laboratory researcher at the NIV who fell ill on 17 April [43]. The entire NIV institute was closed and all of its 200 employees placed in quarantine in a hotel. Subsequent investigation confirmed these first three cases as SARS, and eventually identified a total of nine cases, in three generations, including health care workers and their family contacts44. Neither of the two primary patients had worked with live SARS virus, and WHO investigators had “serious concerns” regarding biosafety procedures at the NIV45.
Several Chinese and international groups investigated the outbreak at the NIV, and identified in retrospect two additional SARS laboratory infections at the NIV that had previously gone unrecognized and had begun in February 2004 [46]. A joint China CDC and WHO investigation found many shortcomings in biosecurity at the NIV, and traced the specific cause of the outbreak to an inadequately inactivated preparation of SARS virus that was used in general (not biosecure) laboratory areas in the NIV, including the one in which the two primary cases worked. It had not been tested to confirm its safety after inactivation, as it should have been. The WHO also found more general shortcomings in the handling of live SARS virus and a lack of surveillance of laboratory personnel for laboratory infections.
Li Liming, director of the China CDC and his deputy directory, the director of the NIV and his deputy director, and the director of the division where the two index cases worked were removed from their positions and found guilty of negligence in overseeing safety at the institution47. The Chinese government also decided to move the China CDC campus from its position in a residential neighborhood to an area “more remote from downtown,” and to allocate funds for more advanced laboratory equipment and infrastructure48.
Interestingly, the virology community is still reticent to discuss laboratory escapes: despite the considerable alarm these escapes created in the public health community and the participation of US CDC personnel in their investigation, they go unmentioned in the “10 years after” historical review of SARS by the CDC.49
Example 5: Foot and Mouth Disease (FMD) from Pirbright 2007
Foot and Mouth Disease (FMD) is a veterinary disease that affects primarily cloven-hoofed domestic animals (pigs, sheep and cattle). It has been eradicated in North America and most of Europe. It is highly transmissible, capable of spreading through direct contact and even through some prepared meats (sausages, airline food), on boots of farm workers (or tourists’ shoes: that’s why there’s that question “have you visited a farm” on the re-entry customs checklist coming into the USA), and even by aerosol spread.
FMD only occasionally causes a mild disease in humans, though exposed humans can carry the virus for up to three days, potentially an important method of spread. FMD causes a more serious disease in animals. Often fatal in young animals, survivors are stunted and lose their economic value. Adult animals die less often, but fail to gain weight or drop in milk production, and can become carriers. Most importantly, strict international quarantine regulations mean that an outbreak will cause all livestock and meat from that country to be banned from international trade. Various methods of outbreak control exist, but all are draconian, requiring massive culling (killing) of “in contact” but otherwise healthy animals surrounding index cases. Restrictions on all animal movement and often all commerce through infected areas are imposed, resulting in secondary economic losses from loss of tourism and general economic activity.
For instance, in the UK in 2001 a FMD outbreak ran from February to October 2001, with travel and export restrictions lasting into 2002. To control the outbreak, all susceptible livestock within 3 kms of an active case were culled. At its peak, 80,000-93,000 animals a week were killed and burned on farms, a total about 10 million sheep and cattle. Its direct cost was about $6.9 billion with overall costs to the British economy estimated at $16 billion.
On August 3, 2007 an outbreak of FMD was reported on a farm in the UK, initially with at least 38 cases in cattle identified. Quarantine measures were introduced and an investigation begun, with culling of surrounding livestock. Most countries banned UK livestock and meat exports. The virus was quickly identified as a strain that had caused a 1967 outbreak in the UK, but was not currently circulating in animals anywhere. Another case of “frozen evolution.” However, this outbreak was 2.8 miles (4.6 kms) south of Pirbright, where the only two facilities in the UK that were authorized to hold FMD virus were located. One was the UK Institute for Animal Health (IAH), the other Merial, a commercial veterinary FMD vaccine manufacturer. They both used the 1967 FMD strain, the Merial facility in large amounts (10,000 l) for vaccine manufacture. Operations were suspended at Merial on August 4 and its license to operate withdrawn. A second FMD outbreak quickly appeared near the first, and animal movement with the UK restricted and quarantine zones encompassing both the Pirbright campus and two affected farms were put in place on Aug 750. An initial investigation also published August 7 found no evidence for aerosol or surface water transmission of FMD virus from Pirbright, was investigating other wastewater issues, and suggested human carriage might have occurred51.
Investigation eventually showed that a waste-water line carrying partially treated waste water from the Merial vaccine plant to the final waste treatment plant run by IAH had gone without routine inspection or maintenance, was damaged, leaking, and had an unsealed manhole opening to the surface, so was capable of contaminating ground and surface water. It became clear that Merial and the IAH each considered the other responsible for such inspection and maintenance, and it had gone undone. The non-secure wastewater line ran through a construction area that recent heavy rains had turned into deep mud, and construction vehicles traversed it and exited the Pirbright campus without inspection or monitoring. These trucks sometimes used the road that passed by the first affected farm. It was concluded that contaminated mud from the defective wastewater line at Pirbright had been carried on tyres or underbody of construction vehicles and caused the first outbreak52.
For a brief time the outbreak was thought to have ended, and restrictions in the Pirbright area were lifted September 8, 2007. The UK applied to the EU to lift most restrictions on animal exports from the UK to EU on September 11, 2007.
However, on September 12, 2007 FMD was again reported, this time 30 miles north of Pirbright, again with the same 1967 strain of FMD. From September 18-30 multiple additional outbreaks of FMD appeared in the same area. A national embargo on all animal shipment was imposed, and new surveillance zones expanded rapidly until, overlapping they encompassed a portion of Heathrow Airport and were cut across by the major M4, M3 and M25 motorways. Rapid (real-time) genomic analysis had been ongoing during this outbreak, and indicated a single escape of FMD from Pirbright, which first spread between the two farms of the August outbreak, then went unnoticed at third farm before it blossomed again in mid September. Follow-up investigations identified the intermediate farm53.
The 2007 UK FMD outbreak identified 278 infected animals, and required 1578 animals to be culled54. It disrupted UK agricultural production and exports, and cost an estimated 200 million pounds. The ban of meat exports was particularly damaging as UK beef had only just exited a 10-year embargo by the EU because of BSE (Mad Cow Disease) in May of 2006.
FMD is such an easily transmitted virus with such potential to cause massive economic damage it would appear that manipulating it in a virology laboratory in a FMD free area is manifestly fraught with hazard. Particularly when it might escape by an “invisible” breach in biosafety as it did at Pirbright, and where it might lurk undetected despite heavy surveillance as it did between the two outbreaks.
In the US, previous law had banned it on the continental US, so FMD virus is currently only held in the USDA Plum Island facility off of Long Island (in a facility originally built in the 1950s for anti-animal BW [biological weapons] work). Currently a replacement facility under the Department of Homeland Security (DHS), the National Bio-and Agro-Defense Facility (NBAF) is under construction in Manhattan, KS. The move of FMD research to the agricultural heartland of the US was opposed by many groups, including the GAO, but DHS decided on the KS location and construction is ongoing. So much for learning from other’s experience.
Conclusions
There are some common themes in these narratives of escaped pathogens. Undetected flaws in the functioning of what was considered at the time to be an adequate standard of technical biocontainment is one theme, as demonstrated in the UK smallpox and FMD cases. Transfer to and handling of inadequately inactivated preparations of dangerous pathogens in areas of the laboratory with reduced biosecurity levels (allowable if the preparation is actually inactivated) is another theme, demonstrated in the SARS and VEE escapes. Poor training of personnel and slack oversight of laboratory procedures negated policy efforts by national and international bodies to achieve biosecurity in the SARS and UK smallpox escapes. The recent appearance of a cohort of immunologically naïve people in the general population, which previously had been uniformly immune was a factor in the UK smallpox and the 1977 H1N1 escapes; in this regard it should be remembered that there is no immunity at all in the general population to most potentially pandemic pathogens currently under discussion, such as Avian influenza and SARS.
It is hardly reassuring that despite stepwise technical improvements in containment facilities and increased policy demands for biosecurity procedures in the handling of dangerous pathogens, that escapes of these pathogens regularly occur and cause outbreaks in the general environment. Looking at the problem pragmatically, question is not if such escapes will happen in the future, but rather what the pathogen may be and how such an escape will be contained, if indeed it can be contained at all.
Advances in genetic manipulation now allow the augmentation of virulence and transmissibility in dangerous pathogens, and such experiments have been funded and performed, notably in the H5N1 avian influenza virus. The advisability of performing such experiments at all, and particularly in laboratories placed at universities in heavily populated urban areas, where laboratory personnel who are potentially exposed are in daily contact with a multitude of susceptible and unaware citizens is clearly in question.
If such manipulations should be allowed at all, it would seem prudent to conduct them in isolated laboratories where personnel are sequestered from the general public and must undergo a period of “exit quarantine” before re-entering civilian life55. Such isolated “detached duty”, while inconvenient for the lifestyle of virologists, is hardly foreign to them, since many experience prolonged periods of inconvenient and dangerous field work in the collection of viruses in the field, and certainly many other natural scientists do prolonged and isolated field work as well. The “inconvenience” barrier that requiring such isolation may present to principal investigators and other personnel may act as a natural screening factor to insure that dangerous manipulations to dangerous pathogens are only undertaken when genuinely indicated.
_______________
REFERENCES
1 Marc Lipsitch and Barry R. Bloom 2012. Rethinking Biosafety in Research on Potential Pandemic Pathogens . mBio 3(5): . doi:10.1128/mBio.00360-12.
2 Klotz, LC. The Human Fatality and Economic Burden of a Man-made Influenza Pandemic: A Risk Assessment. Unpublished Jan 2014. May be accessed at http://armscontrolcenter.org/The_Human_ ... 1-5-14.pdf or http://bio-security.org/wp-content/uplo ... 1-5-14.pdf
3 Merler S, Ajelli M, Fumanelli L, and Vespignani A. Containing the accidental laboratory escape of potential pandemic influenza viruses. BMC Medicine 2013, 11:252 doi:10.1186/1741-7015-11-252 http://www.biomedcentral.com/1741-7015/11/252
4 Scientists call for urgent talks on mutant-flu research in Europe. By Heidi Ledford. Nature 20 December 2013 http://www.nature.com/polopoly_fs/7.145 ... letter.pdf
5 Klotz LC, Sylvester EJ. The unacceptable risks of a man-made pandemic. Bulletin of the Atomic Scientists 7 Aug 2012.
6 Henkel R, Miller T, and Weyant RS. Monitoring Select Agent Theft, Loss and Release Reports in the United States—2004-2010. Applied Biosafety 17 (4) 2012 http://www.absa.org/abj/abj/121704FAHenkel.pdf Escaped Viruses-final 2-17-14
7 Baillie GJ, et al. Evolutionary dynamics of local Pandemic H1N1/2009 Influenza virus lineages revealed by whole-genome analysis. J Virol 86(1):11-18 Jan 2012.
8 Shooter RA. Report of the Investigation into the Cause of the 1978 Birmingham Smallpox Occurrence. HMSO 1980. Available: http://www.official-documents.gov.uk/do ... 8/0668.pdf
9 Fenner F. Henderson D. A. Et al. Smallpox and its Eradication. Published by World Health Organization, Geneva, 1988. ISBN 10: 9241561106 / ISBN 13: 9789241561105 available online: http://apps.who.int/iris/handle/10665/39485.
10 Kung HC, et al. Influenza in China in 1978: recurrence of influenza virus A subtype H1N1. Bull WHO 56(6):913-918 (1978).
11 Beveridge WIB. Where did red flu come from? New Sci. 1978 Mar 23;77/1095:790–1.
12 Nakajima K, Desselberger U, Palese P. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature. 1978;274:334–339.
13 Scholtissek C, von Hoyningen V, Rott R. Genetic relatedness between the new 1977 epidemic strains (H1N1) of influenza and human influenza strains isolated between 1947 and 1957 (H1N1). Virology. 1978;89:613–617
14 Chakraverty P. The return of the historic influenza A H1N1 virus and its impact on the population of the United Kingdom. J Hyg, Camb (1982) 89: 89-100
15 Shortridge KF. Et al. Reappearance of H1N1 influenza virus in man: evidence for the persistence of the virus in domestic chickens. Bull WHO 57(3):475-477 (1979)
16 Zhang G, Shoham D, Gilichinsky D, Davydov S, Castello JD, Rogers SO. Evidence of influenza a virus RNA in siberian lake ice. J Virol. 2006 Dec;80(24):12229-35. Epub 2006 Oct 11. Erratum in: J Virol. 2007 Mar;81(5):2538.
17 Gang Zhang, Dany Shoham, David Gilichinsky, Sergei Davydov, John D. Castello, and Scott O. Rogers. Correction: Evidence of influenza a virus RNA in Siberian lake ice. J Virol. 2007 March; 81(5): 2538. doi: 10.1128/JVI.02773-06 PMCID: PMC1865937
18 Worobey M.J. Phylogenetic evidence against evolutionary stasis and natural abiotic reservoirs of influenza A virus Virol. 2008 Apr;82(7):3769-74. doi: 10.1128/JVI.02207-07. Epub 2008 Jan 30.
19 Zimmer SM, Burke DS. Historical perspective–Emergence of influenza A (H1N1) viruses. N Engl J Med. 2009;361:279–285. [PubMed]
20 Nelson MI, Viboud C, Simonsen L, Bennett RT, Griesemer SB, St George K, Taylor J, Spiro DJ, Sengamalay NA, Ghedin E, Taubenberger JK, Holmes EC. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 2008 Feb 29;4(2):e1000012. doi: 10.1371/journal.ppat.1000012.
21 Wertheim JO. The re-emergence of H1N1 influenza virus in 1977: a cautionary tale for estimating divergence times using biologically unrealistic sampling dates. PLoS One. 2010 Jun 17;5(6):e11184. doi: 10.1371/journal.pone.0011184.
22 Oxford JS. Et al. Naturally occurring temperature-sensitive Influenza A viruses of the H1N1 and H3N2 subtypes. J Gen Virol (1980). 48, 383-389.
23 Scherer WF, et al. Vector incompetency: Its implication in the disappearance of epizootic Venezuelan equine encephalomyelitis virus form Middle America. J Med Etomol 23(1):23-29 1986 Jan 24.
24 Powers AM, et al. Repeated emergence of epidemic/epizootic Venezuelan equine encephalitis fro a single genotype of enzootic subtype ID virus. J Virol 71(9):6697-6705. Sept 1997.
25 Weaver, SC, et al. Genetic evidence for the origins of Venezuelan equine encephalitis virus subtype IAB outbreaks. Am J Trop Med Hyg 60(30): 441-448. 1999.
26 Braulty AC, et al. Potential sources of the 1995 Venezuelan equine encephalitis subtype IC epidemic. J Virol 75(13): 5823-5832 July 2001.
27 Rivas F, et al. Epidemic Venezuelan equine encephalitis in La Guajira, Colombia, 1995. J Infect Dis 1997;175:828-832.
28 Weaver SC, et al. Re-emergence of epidemic Venezuelan equine encephalomyelitis in South America. Lancet 1996 Aug 17 348(9025):346-340.
29 Scherer WF, et al. Vector incompetency: Its implication in the disappearance of epizootic Venezuelan equine encephalomyelitis virus from Middle America. J Med Etomol 23(1):23-29 1986 Jan 24.
30 Braulty AC, et al. Potential sources of the 1995 Venezuelan equine encephalitis subtype IC epidemic. J Virol 75(13): 5823-5832 July 2001.
31 Navarro J-C, et al. Postepizootic persistence of Venezuelan equine encephalitis virus, Venezuela. Emerg Infect Dis 11(12):1907-1915 Dec 2005.
32 Scherer WF, et al. Vector incompetency: Its implication in the disappearance of epizootic Venezuelan equine encephalomyelitis virus from Middle America. J Med Etomol 23(1):23-29 1986 Jan 24.
33 Abraham, T. Twenty-first Century Plague: The Story of SARS. Baltimore MD: Johns Hopkins Univ Press 2005. Is a short, useful summary of the SARS epidemic.
34 International Dimensions of Ethics Education in Science and Engineering: Case Study Series. IDEESE: Cases – Reporting Incidence of SARS: Appendix A. available at: http://www.umass.edu/sts/ethics/sars.html.
35 Zhuaang Shen, et al. Superspreading SARS events, Beijing, 2003 Emerg Infect Dis 2004 10(2):256-260. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3322930/#R1
36 MMWR. Update: Outbreak of Severe Acute Respiratory Syndrome – Worldwide, 2003. MMWR March 28, 2003 / 52(12);241-248. http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5212a1.htm
37 WHO. World Health Report 2007 Ch 3: (illustration): Probable SARS transmission on flight CA112 in March 2003. http://www.who.int/whr/2007/media_centr ... g01_en.pdf
38 Singapore Ministry of Health. Biosafety and SARS Incident in Singapore September 2003. Websource: http://www.biosafety.be/CU/PDF/Report_S ... gapore.pdf
39 WHO Global Alert and Response: Severe Acute Respiratory Syndrome (SARS) in Taiwan, China. 17 Dec 2003. http://www.who.int/csr/don/2003_12_17/en/
40 Reuters: Taipei, Dec 20,2003. Taiwan says scientist likely got SARS in lab slip. Accessed at: http://www.royalsociety.org.nz/2003/12/ ... -taiwan-2/
41 Wu, W C et al. Development of Laboratory Biosafety Management: The Taiwan Experience. Applied Biosafety 12(1):18-25 2007. Accessed at: http://www.absa.org/abj/abj/071201wu.pdf
42 WHO. WHO post-outbreak biosafety guidelines for handling of SARS-CoV specimens and cultures. 18 Dec 2003. Accessible at: http://www.who.int/csr/sars/biosafety2003_12_18/en/
43 WHO Global Alert and Response (GAR). China reports additional SARS cases – update 23 April 2004. http://www.who.int/csr/don/2004_04_23/en/index.html
44 WHO Global Alert and Response (GAR). China confirms SARS in infection in another previously reported case; summary of cases to date – update 5. 30 April 2004. http://www.who.int/csr/don/2004_04_30/en/index.html
45 WHO Global Alert and Response (GAR). China’s latest SARS outbreak has been contained, but biosafety concerns remain – update 7. 18May2004. http://www.who.int/csr/don/2004_05_18a/en/index.html
46 Zhao Xiaojian, Zhu Xiaochao. China CDC Blamed for SARS escape. Caijing.com.cn Daily Update May 20, 2004. http://english.caijing.com.cn/2004-05-20/100013918.html
47 SCiDevNet website. Top Chinese Scientists ‘punished’ over SARS outbreak. July 8, 2004. http://www.scidev.net/global/health/new ... utbre.html
48 Zhang Feng (China Daily). Officials punished for SARS virus leak. Chinadaily.com.cn July 7, 2004. http://www.chinadaily.com.cn/english/do ... 344755.htm
49 CDC website. Remembering SARS: A deadly puzzle and the efforts to solve it. Online source: http://www.cdc.gov/about/history/sars/feature.htm
50 DEFRA. Declaration of a Protection Zone and Surveillance Zone 7Aug2007. http://webarchive.nationalarchives.gov. ... rz0807.pdf
51 HSE. Initial report on potential breaches of biosecurity at the Pirbright site 2007. Web source: http://webarchive.nationalarchives.gov. ... 0807b.htm/
52 Logan, P. Final report on potential breaches of biosecurity at the Pirbright site 2007. Health and Safety Executive September 2007. http://www.hse.gov.uk/news/2007/finalreport.pdf
53 Cottam EM, et al. Transmission pathways of Food-and-Mouth Disease Virus in the United Kingdom in 2007. PLosPathogens April 2008 4(4) e1000050. Web: http://www.plospathogens.org/article/in ... at.1000050
54 Juleff, N. The 2007 UK FMD outbreak: field investigation perspective. Institute for Animal Health (UK). PowerPoint presentation. Available online: http://www.fao.org/ag/againfo/commissio ... ctives.pdf
55 Klotz, LC and Sylvester, E. The unacceptable risks of a man-made pandemic. Bulletin of the Atomic Scientists. 7 August 2012. http://thebulletin.org/unacceptable-ris ... e-pandemic
- admin
- Site Admin
- Posts: 36135
- Joined: Thu Aug 01, 2013 5:21 am
Re: U.S. government gave $3.7 million grant to Wuhan lab at
The Evidence which Suggests that This Is No Naturally Evolved Virus: A Reconstructed Historical Aetiology of the SARS-CoV-2 Spike
by Birger Sørensen, Angus Dalgleish & Andres Susrud
Immunor & St Georges University of London
July 1, 2020
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
ABSTRACT
To discover exactly how to attack SARS-CoV-2 safely and efficiently, our vaccine candidate Biovacc-19 was designed by first carefully analysing the biochemistry of the Spike. We ascertained that it is highly unusual in several respects, unlike any other CoV in its clade. The SARS-CoV-2 general mode of action is as a co-receptor dependent phagocyte. But data shows that simultaneously it is capable of binding to ACE2 receptors in its receptor binding domain. In short, SARS-CoV-2 is possessed of dual action capability. In this paper we argue that the likelihood of this being the result of natural processes is very small. The spike has six inserts which are unique fingerprints with five salient features indicative of purposive manipulation. We then add to the bio-chemistry a diachronic dimension by analysing a sequence of four linked published research projects which, we suggest, show by deduction how, where, when and by whom the SARS-CoV-2 Spike acquired its special characteristics. This reconstructed historical aetiology meets the criteria of means, timing, agent and place to produce sufficient confidence to reverse the burden of proof. Henceforth, those who would maintain that the Covid-19 pandemic arose from zoonotic transfer need to explain precisely why this more parsimonious account is wrong before asserting that their evidence is persuasive, most especially when, as we also show, there are puzzling errors in their use of evidence.
Introduction: Why does this matter?
No-one has ever produced a safe and effective vaccine against a coronavirus. In the context of a forthcoming paper addressing contingency actions cognizant of this fact, the potentialities for 'trained immunity' from 'new old friends' in the form of Bacillus Calmette–Guérin (BCG), Microbacillus vaccae (IMM-102) and most especially Microbacillus obuense (IMM-101) by stimulating the innate immune system and especially Delta Gamma T cells are explored; and a salutary review of failed vaccine programmes is included (Kleen et al., 2020). On 28th April 2020, Nature published a graphical guide to eight conceptual approaches featuring in current explorations of around 90 vaccine development programmes intended to counter SARS-CoV-2 (Callaway, 2020).
We have just (2nd June 2020) published Biovacc-19 in QRB-Discovery: a candidate vaccine for this daunting task (Sørensen et al., 2020). Its mode of action is unique and therefore is not included in the Nature review. In our paper we gave reasons why the virus vector or RNA vector based approaches that are the basis of the eight methodologies reviewed in Nature are unlikely to prove immunogenic and why either, but especially RNA vectored models, may carry significant risk of Antibody Dependent Enhancement (ADE). As we have detailed in QRB-D, we have seen such a story before over thirty years in the failure of all three mainstream vaccine approaches to HIV, which we predicted but were disbelieved.
As with our HIV vaccine, the methodology underpinning Biovacc-19 first analysed fully the virus target. In this case we published the general mode of action for infectivity of SARS-CoV-2. Doing this took us into a fundamental exploration of the biochemistry and structure of the SARS-CoV-2 Spike which is highly singular, possessed of features that we have not seen before and which are not present in other SARS viruses of that clade. We posited that the SARS-CoV-2 general mode of action is as a co-receptor dependent phagocyte. But unusually, simultaneously, data shows that it is capable of binding to ACE2 receptors in its receptor binding domain. In short, SARS CoV-2 is possessed of dual action capability. How do we think this was made possible? That is the subject of this paper. We shall argue from evidence below that the likelihood of this being the result of natural processes is very small.
The co-receptor dependent phagocytic general method of action for infectivity and pathogenicity of SARS-CoV-2 appears to be specifically related to cumulative charge resulting from inserts placed on the surface of the Spike receptor binding domain, right next to the receptor binding motif. That SARS-CoV-2 has charged inserts is not in dispute (Zhou et al., 2020) What we have shown that is new is that the SARS-CoV-2 Spike carries significant additional charge (isoelectric point (pI) pI=8.2) compared to human SARS-CoV Spike,( pI = 5.67) and the implications thereof. Basic domains -- partly inserted, partly substituted amino acids and partly redistributed from outside the receptor binding domain -- explain the salt bridges formed between the SARS-CoV-2 Spike and its co-receptors on the cell membrane. We comment further on the significance of this in the next section.
Puzzling features
An influential paper was published in Nature Medicine on 17 March 2020. Andersen et al observed that several mutations have occurred in the receptor binding domain of SARS-CoV-2. These, they suggested, therefore sustain an hypothesis of natural evolution (Andersen et al., 2020). We do not agree. We do agree that it is indeed correct that several such mutations are to be seen and in a forthcoming companion article to this one, about three other viruses of interest, we will discuss further Andersen et al's evidence and argumentation in that context. But here we observe only that the contention that it is improbable that Covid-19 emerged through laboratory manipulation of a related SARS-CoV-like coronavirus because the ACE2 binding is not ideal is weakened because Andersen et al cite two authorities which actually say the reverse of what they say that they say.
Wan et al are cited by Andersen et al but offer them no support (Wan et al., 2020). Wan et al say, correctly in our view, that computational structural modelling of complex virus-receptor interactions can be used for structural predictions and that such models can potentially be used for Gain-Of-Function modelling. It is well known that models have been developed from data generated in animal model systems such as the palm civet. Wan et al say that the SARS-CoV-2 binding to the ACE2 receptor confirms the accuracy of the structural predictions. Therefore the data and conclusion in Wan et al contradicts Andersen et al's opinion that it is improbable that the virus could have emerged through laboratory manipulation.
There is a similar problem with (Sheahan et al., 2008). This deals with research on a civet strain SZ16 and the infective strain SARS-CoV Urbani. These strains were used to create a chimeric virus icSZ16-S. Sheahan et al go on to explain that by in vitro evolution of the chimeric virus icSZ16-S on human airway epithelial (HAE) cells in the lab, they have been able to produce two new viruses binding to such HAE cells. Therefore this reference supports the very opposite of the Andersen et al hypothesis. We are immediately wary of any paper containing such egregious errors.
Our discovery of the high pI number, the high accumulated charge and how it comes about, in the course of our bio-chemical analysis, suggested several features which individually seem unlikely to be the result of natural evolution and which, taken together, and applying Occam's Razor to hone the most parsimonious hypothesis, make natural evolution a less likely explanation than purposive manipulation, specifically for Gain of Function.
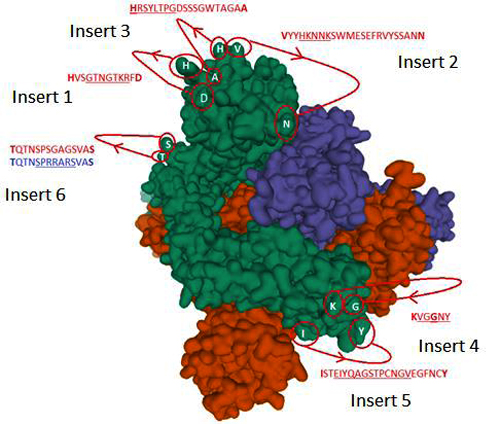
Figure 1: The identified inserts examined in the PDB 6VXX electron microscopy structure (Walls et al., 2020) The sequences highlighted in red could not be found in the cryo-electron microscopy structure data. The 6 aligned sequences in Fig. 1 in (Sørensen et al.,2020) are underlined in the missing sequences. Bold amino acids indicate first and last amino acids used to build the structure where the missing part is in between. Insert 6 did not have the same sequence in 6VXX as in the reference Sars-CoV-2 sequence. The authors stated that a designed mutated strain lacking the furin cleavage site residues was used.
Figure 2: The positive charged domains associated with cysteine loops Cys131-Cys166, Cys336-Cys361, Cys391-Cys525, Cys538-Cys590. As can be seen, there is a high concentration of positive charged surface exposed amino acids within the receptor domain next to the receptor binding motif at the top of the spike. The location of the positive charged amino acids in red circles on the right-hand side of the figure points out their surface exposure making them available for cell attachment as discussed in (5) below. Insert 2 (HKNNK) in Figure 1 above is located within the Cys131-Cys166 loop but was omitted in the Cryo-EM structure shown in dashed lines (Walls et al, 2020). However, charged amino acids belong to the hydrophilic group of amino acids and are most likely surface exposed.
To recapitulate Fig 2 from our vaccine paper, there are 6 inserts which make the SARS-CoV-2 Spike structurally special. They are unique fingerprints of the SARS-CoV-2 Spike which deserve to be highlighted in support of this view; and there are five salient features that strengthen the case for purposive manipulation in the laboratory.
Taken all together, we suggest that our research findings on the general mode of action for infectivity of SARS-CoV-2 and the further puzzling features just mentioned, justify the question of the historical aetiology of these manipulations.
We did not need to address this issue diachronically for the purposes of vaccine design. However, it is important for a soundly based understanding of the present and potential future epidemiology of the Covid-19 pandemic and for strategies for its management. Therefore to our earlier amino-acid level of biochemical analysis we now add here a forensic analysis of published research literature concerning SARS-CoV-2. We will extend this type of analysis to three other viruses in the companion article.
Since, regrettably, international access has not been allowed to the relevant laboratories or materials, since Chinese scientists who wished to share their knowledge have not been able to do so and indeed since it appears that preserved virus material and related information have been destroyed, we are compelled to apply deduction to the published scientific literature, informed by our own biochemical analyses. We refute pre-emptively objection that this methodology does not result in absolute proof by observing that to make such a statement is to misunderstand scientific logic. The longer the chain of causation of individual findings that is shown, especially converging from different disciplines, the greater the confidence in the whole. We posit that the evidence below attains a high level of confidence.
A sequence of four linked research papers is explained
A comprehensive review of the relevant literature shows that a substantial amount of directly relevant gain-of function research has been undertaken. Four studies are especially noteworthy. They are linked in two ways: scientifically, in that the third and fourth build upon the results of the first and second, and in continuity of the institution and personnel across all four. The Wuhan Institute of Virology is a key collaborator in all these projects and Dr Zheng-Li Shi is one of the Institute's most experienced virologists and bat specialists. She is a common thread through all the key research projects.
Now recollect that Menachery V.D et al in 2015 had shown that their chimeric virus SHC014-MA15 could, against their prediction, very successfully infect primary human upper airway epithelial cells (HAE) from the cell-line 2B4 Calu-3. With this in mind, we next observed that in the Covid-19 pandemic, a well-reported symptom in the early phase of the infection is loss of taste, headache and a sore throat. We have discussed this issue in the QRBD article in detail. But to summarise: in 2015 in a research review (Workman et al, 2015) discussed bitter/sweet taste receptors and the role these receptors play in mediating airway immune functions. They concluded thus: "Over the past several years, taste receptors have emerged as key players in the regulation of innate immune defenses in the mammalian respiratory tract. Several cell types in the airway, including ciliated epithelial cells, solitary chemosensory cells, and bronchial smooth muscle cells, all display chemoresponsive properties that utilize taste receptors."
Therefore we hypothesise the reconstructed historical aetiology of the Spike as follows:
In 2008, Dr Zheng-Li Si and WIV colleagues successfully demonstrated technical capabilities to interchange RBD’s between bat SARS-like and human SARS viruses. Building upon this, the 2010 work (Hou et al, 2010) perfected the ability to express receptors on human cells. On these foundations, the central Gain of Function work that underpins the functionalities of SARS-CoV-2 took place, carrying the WIV spike and plasmid materials to bond successfully to a UNC Chapel Hill human epithelial cell-line. This work (Menachery et al) produced a highly infectious chimeric virus optimised to the human upper respiratory tract. In convergent support of this hypothesis, both Lu (Lu et al, 2020) and Jia (Jia et al, 2020) have now, in January and April 2020, shown that SARS-CoV-2 has a bat SARS-like backbone but is carrying an RBD from a human SARS and Zhan et al have, like us, noted unusual adaption to humans from the first isolate. In the 2015 Chapel Hill work it was only ACE2 receptors that were discussed. However, in 2018 Zhou P. et al demonstrated capabilities to clone other receptors like APN and DPP4 and to test and compare these against the (intestine) tissue specific SADS-CoV identified. Then, in the 2019-20 Covid-19 pandemic, profuse symptoms indicating compromise of the bitter/sweet receptors are reported. Taken all together, this implies that by employing insights gained after 2015, as just deduced, a further optimization of the 2015 chimeric virus for additional binding to receptors/co-receptors such as bitter/sweet specific upper airway epithelia receptors occurred. That would help to explain the otherwise puzzling high infectivity and pathology associated with SARS-CoV-2 and hence also help to explain the social epidemiology of its spread.
Conclusion
We have deduced the internal logic of published research which resulted in the exact functionalities of SARS-CoV-2, including the convergence of agreement from difference classes of source, the timings of the stages of the research and the development of documented capabilities by named institutions and individuals. These meet the criteria of means, timing, agent and place in this reconstructed historical aetiology to produce sufficient confidence in the account to reverse the burden of proof. Henceforth, those who would maintain that the Covid-19 pandemic arose from zoonotic transfer need to explain precisely why this more parsimonious account is wrong before asserting that their evidence is persuasive, most especially when, as we have indicated, we note puzzling errors in their use of evidence. In our companion article, in a similar forensic manner we will explore the primary evidence used to sustain the hypothesis of zoonotic transfer. In neither this article nor the next do we speculate about motive.
Oslo & London 1 July 2020
_______________
References:
Alan D. Workman, James N. Palmer, Nithin D. Adappa and Noam A. Cohen. "The Role of Bitter and Sweet Taste Receptors in Upper Airway Immunity " Curr Allergy Asthma Rep. 2015 December ; 15(12): 72. doi:10.1007/s11882-015-0571-8.
Andersen, K. G., Rambaut, A., Lipkin, W.I., Holms, E. C., Garry, R. F., 2020, "The proximal origin of SARS-CoV-2". Nat Med. https://doi.org/10.1038/s41591-020-0820-9
Callaway E, "The Race for Coronavirus Vaccines", Nature, Vol 580, 30 April 2020, pp 576-7 (https://www.nature.com/articles/d41586-020-01221-y)
Hou Y, Peng C, Yu M, Li Y, Han Z, Li F, Wang LF, Shi Z., 2010, "Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry", Arch Virol, 155:1563–1569, doi: 10.1007/s00705-010-0729-6Hou 2010
Thomas-Oliver Kleen, Alicia A Galdon, Andrew S Macdonald and Angus G Dalgleish, Mitigating dysfunctional immunity in COVID-19: Trained Immunity, BCG and ‘new old friends’, 2020. (paper submitted)
Lin Xu, Fuqiang Zhang, Weihong Yang, Tinglei Jiang, Guanjun Lu, Biao He, Xingyu Li, Tingsong Hu, Gang Chen, Yun Feng, Yuzhen Zhang, Quanshui Fan, Jiang Feng, Hailin Zhang & Changchun Tu, 2016, "Detection and characterization of diverse alpha- and betacoronaviruses from bats in China", Virologica Sinica, 31 (1): 69–77, doi: 10.1007/s12250-016-3727-3
Marzi A, Gramberg T, Simmons G, Möller P, Rennekamp AJ, Krumbiegel M, Geier M, Eisemann J, Turza N, Saunier B, Steinkasserer A, Becker S, Bates P, Hofmann H, Pöhlmann S, (2004) "DC-SIGN and DC-SIGNR Interact with the Glycoprotein of Marburg Virus and the S Protein of Severe Acute Respiratory Syndrome Coronavirus", Journal of Virology, DOI: 10.1128/JVI.78.21.12090-12095.
Roujian Lu, Xiang Zhao, Juan Li, Peihua Niu, Bo Yang, Honglong Wu, Wenling Wang, Hao Song, Baoying Huang, Na Zhu, Yuhai Bi, Xuejun Ma, Faxian Zhan, Liang Wang, Tao Hu, Hong Zhou, Zhenhong Hu, Weimin Zhou, Li Zhao, Jing Chen, Yao Meng, Ji Wang, Yang Lin, Jianying Yuan, Zhihao Xie, Jinmin Ma, William J Liu, Dayan Wang, Wenbo Xu, Edward C Holmes, George F Gao, Guizhen Wu, Weijun Chen, Weifeng Shi, Wenjie Tan. “Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding”. Lancet 2020; 395: 565–74. https://doi.org/10.1016/ S0140-6736(20)30251-8
Sørensen, B., Susrud, A., & Dalgleish, A. (2020). "Biovacc-19: A Candidate Vaccine for Covid-19 (SARS-CoV-2) Developed from Analysis of its General Method of Action for Infectivity". QRB Discovery, 1-17. doi:10.1017/qrd.2020.8
Timothy Sheahan, Barry Rockx, Eric Donaldson, Amy Sims, Raymond Pickles, Davide Corti, Ralph Baric, 2008, “Mechanisms of Zoonotic Severe Acute Respiratory Syndrome Coronavirus Host Range Expansion in Human Airway Epithelium”. J Virol. https://doi.org/10.1128/JVI.02041-07
Vineet D Menachery, Boyd L Yount Jr, Kari Debbink, Sudhakar Agnihothram, Lisa E Gralinski, Jessica A Plante, Rachel L Graham, Trevor Scobey, Xing-Yi Ge, Eric F Donaldson, Scott H Randell, Antonio Lanzavecchia, Wayne A Marasco, Zhengli-Li Shi & Ralph S Baric , 2015," A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence", Nature Medicine, 21(12): 1508–1513. doi:10.1038/nm.3985
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D, (2020) “Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein”, Cell, Volume 181, Issue 2, 16 April, Pages 281-292.e6, DOI: https://doi.org/10.1016/j.cell.2020.02.058
Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. "Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus". J Virol 94: e00127-20. https://doi.org/10.1128/JVI.00127-20.
Wuze Ren, Xiuxia Qu, Wendong Li, Zhenggang Han, Meng Yu, Peng Zhou, Shu-Yi Zhang, Lin-Fa Wang, Hongkui Deng and Zhengli Shi, 2008, “Difference in Receptor Usage between Severe Acute Respiratory Syndrome (SARS) Coronavirus and SARS-Like Coronavirus of Bat Origin”. J Virol. https://doi.org/10.1128/JVI.01085-07
Xian-Dan Lin, Wen Wanga, Zong-Yu Hao, Zhao-Xiao Wang, Wen-Ping Guo, Xiao-Qing Guan, Miao-Ruo Wang, Hong-Wei Wang, Run-Hong Zhou, Ming-Hui Li, Guang-Peng Tang, Jun Wu, Edward C. Holmes, Yong-Zhen Zhang, 2017, "Extensive diversity of coronaviruses in bats from China", Virology 507, 1–10, https://doi.org/10.1016/j.virol.2017.03.019
Yong Jia, Gangxu Shen, Yujuan Zhang, Keng-Shiang Huang, Hsing-Ying Ho, Wei-Shio Hor, Chih-Hui Yang, Chengdao Li, Wei-Lung Wang. “Analysis of the mutation dynamics of SARS-CoV-2 reveals the spread history and emergence of RBD mutant with lower ACE2 binding affinity”. bioRxiv April 11,2020. https://doi.org/10.1101/2020.04.09.034942
Zhan S.H. et.al. (2020) "SARS-CoV-2 is well adapted for humans. What does this mean for re-emergence"- bioRxiv
Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XLL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG & Ma JY, (2018) "Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin", Nature 556, 255–258, https://doi.org/10.1038/s41586-018-0010-9
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF & Shi ZL, (2020) "A pneumonia outbreak associated with a new coronavirus of probable bat origin", Nature 579, 270–273, https://doi.org/10.1038/s41586-020-2012-7
by Birger Sørensen, Angus Dalgleish & Andres Susrud
Immunor & St Georges University of London
July 1, 2020
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
YOU ARE REQUIRED TO READ THE COPYRIGHT NOTICE AT THIS LINK BEFORE YOU READ THE FOLLOWING WORK, THAT IS AVAILABLE SOLELY FOR PRIVATE STUDY, SCHOLARSHIP OR RESEARCH PURSUANT TO 17 U.S.C. SECTION 107 AND 108. IN THE EVENT THAT THE LIBRARY DETERMINES THAT UNLAWFUL COPYING OF THIS WORK HAS OCCURRED, THE LIBRARY HAS THE RIGHT TO BLOCK THE I.P. ADDRESS AT WHICH THE UNLAWFUL COPYING APPEARED TO HAVE OCCURRED. THANK YOU FOR RESPECTING THE RIGHTS OF COPYRIGHT OWNERS.
Therefore we hypothesise the reconstructed historical aetiology of the Spike as follows:
In 2008, Dr Zheng-Li Si and WIV colleagues successfully demonstrated technical capabilities to interchange RBD’s between bat SARS-like and human SARS viruses. Building upon this, the 2010 work (Hou et al, 2010) perfected the ability to express receptors on human cells. On these foundations, the central Gain of Function work that underpins the functionalities of SARS-CoV-2 took place, carrying the WIV spike and plasmid materials to bond successfully to a UNC Chapel Hill human epithelial cell-line. This work (Menachery et al) produced a highly infectious chimeric virus optimised to the human upper respiratory tract. In convergent support of this hypothesis, both Lu (Lu et al, 2020) and Jia (Jia et al, 2020) have now, in January and April 2020, shown that SARS-CoV-2 has a bat SARS-like backbone but is carrying an RBD from a human SARS and Zhan et al have, like us, noted unusual adaption to humans from the first isolate. In the 2015 Chapel Hill work it was only ACE2 receptors that were discussed. However, in 2018 Zhou P. et al demonstrated capabilities to clone other receptors like APN and DPP4 and to test and compare these against the (intestine) tissue specific SADS-CoV identified. Then, in the 2019-20 Covid-19 pandemic, profuse symptoms indicating compromise of the bitter/sweet receptors are reported. Taken all together, this implies that by employing insights gained after 2015, as just deduced, a further optimization of the 2015 chimeric virus for additional binding to receptors/co-receptors such as bitter/sweet specific upper airway epithelia receptors occurred. That would help to explain the otherwise puzzling high infectivity and pathology associated with SARS-CoV-2 and hence also help to explain the social epidemiology of its spread.
-- The Evidence which Suggests that This Is No Naturally Evolved Virus: A Reconstructed Historical Aetiology of the SARS-CoV-2 Spike, by Birger Sørensen, Angus Dalgleish & Andres Susrud
ABSTRACT
To discover exactly how to attack SARS-CoV-2 safely and efficiently, our vaccine candidate Biovacc-19 was designed by first carefully analysing the biochemistry of the Spike. We ascertained that it is highly unusual in several respects, unlike any other CoV in its clade. The SARS-CoV-2 general mode of action is as a co-receptor dependent phagocyte. But data shows that simultaneously it is capable of binding to ACE2 receptors in its receptor binding domain. In short, SARS-CoV-2 is possessed of dual action capability. In this paper we argue that the likelihood of this being the result of natural processes is very small. The spike has six inserts which are unique fingerprints with five salient features indicative of purposive manipulation. We then add to the bio-chemistry a diachronic dimension by analysing a sequence of four linked published research projects which, we suggest, show by deduction how, where, when and by whom the SARS-CoV-2 Spike acquired its special characteristics. This reconstructed historical aetiology meets the criteria of means, timing, agent and place to produce sufficient confidence to reverse the burden of proof. Henceforth, those who would maintain that the Covid-19 pandemic arose from zoonotic transfer need to explain precisely why this more parsimonious account is wrong before asserting that their evidence is persuasive, most especially when, as we also show, there are puzzling errors in their use of evidence.
Introduction: Why does this matter?
No-one has ever produced a safe and effective vaccine against a coronavirus. In the context of a forthcoming paper addressing contingency actions cognizant of this fact, the potentialities for 'trained immunity' from 'new old friends' in the form of Bacillus Calmette–Guérin (BCG), Microbacillus vaccae (IMM-102) and most especially Microbacillus obuense (IMM-101) by stimulating the innate immune system and especially Delta Gamma T cells are explored; and a salutary review of failed vaccine programmes is included (Kleen et al., 2020). On 28th April 2020, Nature published a graphical guide to eight conceptual approaches featuring in current explorations of around 90 vaccine development programmes intended to counter SARS-CoV-2 (Callaway, 2020).
We have just (2nd June 2020) published Biovacc-19 in QRB-Discovery: a candidate vaccine for this daunting task (Sørensen et al., 2020). Its mode of action is unique and therefore is not included in the Nature review. In our paper we gave reasons why the virus vector or RNA vector based approaches that are the basis of the eight methodologies reviewed in Nature are unlikely to prove immunogenic and why either, but especially RNA vectored models, may carry significant risk of Antibody Dependent Enhancement (ADE). As we have detailed in QRB-D, we have seen such a story before over thirty years in the failure of all three mainstream vaccine approaches to HIV, which we predicted but were disbelieved.
As with our HIV vaccine, the methodology underpinning Biovacc-19 first analysed fully the virus target. In this case we published the general mode of action for infectivity of SARS-CoV-2. Doing this took us into a fundamental exploration of the biochemistry and structure of the SARS-CoV-2 Spike which is highly singular, possessed of features that we have not seen before and which are not present in other SARS viruses of that clade. We posited that the SARS-CoV-2 general mode of action is as a co-receptor dependent phagocyte. But unusually, simultaneously, data shows that it is capable of binding to ACE2 receptors in its receptor binding domain. In short, SARS CoV-2 is possessed of dual action capability. How do we think this was made possible? That is the subject of this paper. We shall argue from evidence below that the likelihood of this being the result of natural processes is very small.
The co-receptor dependent phagocytic general method of action for infectivity and pathogenicity of SARS-CoV-2 appears to be specifically related to cumulative charge resulting from inserts placed on the surface of the Spike receptor binding domain, right next to the receptor binding motif. That SARS-CoV-2 has charged inserts is not in dispute (Zhou et al., 2020) What we have shown that is new is that the SARS-CoV-2 Spike carries significant additional charge (isoelectric point (pI) pI=8.2) compared to human SARS-CoV Spike,( pI = 5.67) and the implications thereof. Basic domains -- partly inserted, partly substituted amino acids and partly redistributed from outside the receptor binding domain -- explain the salt bridges formed between the SARS-CoV-2 Spike and its co-receptors on the cell membrane. We comment further on the significance of this in the next section.
Puzzling features
An influential paper was published in Nature Medicine on 17 March 2020. Andersen et al observed that several mutations have occurred in the receptor binding domain of SARS-CoV-2. These, they suggested, therefore sustain an hypothesis of natural evolution (Andersen et al., 2020). We do not agree. We do agree that it is indeed correct that several such mutations are to be seen and in a forthcoming companion article to this one, about three other viruses of interest, we will discuss further Andersen et al's evidence and argumentation in that context. But here we observe only that the contention that it is improbable that Covid-19 emerged through laboratory manipulation of a related SARS-CoV-like coronavirus because the ACE2 binding is not ideal is weakened because Andersen et al cite two authorities which actually say the reverse of what they say that they say.
Wan et al are cited by Andersen et al but offer them no support (Wan et al., 2020). Wan et al say, correctly in our view, that computational structural modelling of complex virus-receptor interactions can be used for structural predictions and that such models can potentially be used for Gain-Of-Function modelling. It is well known that models have been developed from data generated in animal model systems such as the palm civet. Wan et al say that the SARS-CoV-2 binding to the ACE2 receptor confirms the accuracy of the structural predictions. Therefore the data and conclusion in Wan et al contradicts Andersen et al's opinion that it is improbable that the virus could have emerged through laboratory manipulation.
There is a similar problem with (Sheahan et al., 2008). This deals with research on a civet strain SZ16 and the infective strain SARS-CoV Urbani. These strains were used to create a chimeric virus icSZ16-S. Sheahan et al go on to explain that by in vitro evolution of the chimeric virus icSZ16-S on human airway epithelial (HAE) cells in the lab, they have been able to produce two new viruses binding to such HAE cells. Therefore this reference supports the very opposite of the Andersen et al hypothesis. We are immediately wary of any paper containing such egregious errors.
Our discovery of the high pI number, the high accumulated charge and how it comes about, in the course of our bio-chemical analysis, suggested several features which individually seem unlikely to be the result of natural evolution and which, taken together, and applying Occam's Razor to hone the most parsimonious hypothesis, make natural evolution a less likely explanation than purposive manipulation, specifically for Gain of Function.

Figure 1: The identified inserts examined in the PDB 6VXX electron microscopy structure (Walls et al., 2020) The sequences highlighted in red could not be found in the cryo-electron microscopy structure data. The 6 aligned sequences in Fig. 1 in (Sørensen et al.,2020) are underlined in the missing sequences. Bold amino acids indicate first and last amino acids used to build the structure where the missing part is in between. Insert 6 did not have the same sequence in 6VXX as in the reference Sars-CoV-2 sequence. The authors stated that a designed mutated strain lacking the furin cleavage site residues was used.

Figure 2: The positive charged domains associated with cysteine loops Cys131-Cys166, Cys336-Cys361, Cys391-Cys525, Cys538-Cys590. As can be seen, there is a high concentration of positive charged surface exposed amino acids within the receptor domain next to the receptor binding motif at the top of the spike. The location of the positive charged amino acids in red circles on the right-hand side of the figure points out their surface exposure making them available for cell attachment as discussed in (5) below. Insert 2 (HKNNK) in Figure 1 above is located within the Cys131-Cys166 loop but was omitted in the Cryo-EM structure shown in dashed lines (Walls et al, 2020). However, charged amino acids belong to the hydrophilic group of amino acids and are most likely surface exposed.
To recapitulate Fig 2 from our vaccine paper, there are 6 inserts which make the SARS-CoV-2 Spike structurally special. They are unique fingerprints of the SARS-CoV-2 Spike which deserve to be highlighted in support of this view; and there are five salient features that strengthen the case for purposive manipulation in the laboratory.
1. A major part of the spike protein has human-like domains with matured transmission adaption. Blasting the Spike protein with a rolling window of 6 amino acids showed that 78.4% of 6 amino acid windows are human like. This means that with nearly 80% of the spike protein has a built-in stealth property by having high human similarity. Therefore, it is remarkably well-adapted virus for human co-existence. Such high human similarity also implies a high risk for the development of severe adverse events/toxicity and even Antibody Dependent Enhancement (ADE) unless specific precautions are taken when using the Spike protein in any vaccine candidate: precautions that might not suggest themselves to designers employing conventional methodologies and innocent assumptions about the target virus, lacking our detailed anatomisation of it. Furthermore and significantly, Zhan et al also note that, surprisingly, this characteristic is present from the very first isolate (Zhan et al, 2020). This is something that does not sit well with an hypothesis of natural evolution.
2. The Spike displays new amino acid inserts with condensed cumulative charge, all of which are surface exposed (please refer to the reproduced figure from the vaccine paper, above). This is a most significant finding as we mentioned in opening. Being physically located on the surface of the Spike protein greatly increases the infectivity and pathogenicity of the virus, enabling these inserts to participate in binding to co-receptors/negatively charged attachment receptors or even, as we have discovered, to the negatively charged phospholipid heads on the cell membrane. Such a result is typically the objective of gain of function experiments to create chimeric viruses of high potency. Therefore this is a strong indicator of manipulation.
3. The concentration of positive charge is on the receptor binding domain near the receptor binding motif at the top of the Spike protein. As with (2) this is more elegantly explained by an hypothesis of purposive manipulation than one of natural evolution. As can be seen in Figure 2 (side view) of the Spike trimer, the majority of the positive charged amino acids are located near or on the top of the spike protein giving the receptor binding domain a pI=8.906, while the Cov-2 specific Cys538-Cys590 bridge brings in additional charge from 526-560 (with even higher pI=10.03) via the Cys391-Cys525 to positions right next to the receptor binding motif (where the ACE2 receptor is located). It is this which facilitates the dual mode capability, allowing binding to ACE2 and/or to co-receptors/attachments receptors. We posit that such ACE2 independent attachment and infectivity is happening and is evidenced clinically by the Covid-19 disease pattern. It is also reported by Zhou et al (2018). The receptors that are the most likely to be involved are CLEC4M/DC-SIGN (CD209) – see discussion point (5) below.
4. The Spike is so configured that it can bind to cell tissue without use of the ACE2 receptor. Clinically it is widely observed that the Covid-19 virus compromises the functions of olfaction and bitter/sweet receptors, erythrocytes, t-cells, neurons and various tissues such as intestine epithelia. These different targets do not engage and use ACE2 receptor binding. The concentration of high positive charge in and around the top of the Spike protein and the potential to use opposite charged attachment-/co-receptors can facilitate binding and infection in the general mode of action for infectivity that we published in detail in QRBD. In 2018 Zhou P et.al. 2018 found that a new Corona virus which they named SADS (Swine Acute Diarrhoea Syndrome) could infect the intestine and kill piglets without use of ACE2, aminopeptidase N (APN) or dipeptidyl peptidase 4 (DPP4) receptors.[9] We have done a blast analysis of the SADS Spike S1 protein and could find no trace of ACE2 RBM. The significance of this will become clear in the next point and the next section.
5. Location and concentration of charge on the attachment receptor CLEC4M/DC-SIGN (C-type Lectin domain family 4 member M (CLEC4M)/ Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin(DC-SIGNR) also known as CD209) (Marzi et al., 2004). Analysis of the CLEC4M attachment receptor shows an overall pI=5.23 where the C-type lectin tail 274-390 has a pI=4.4. However, due to the two disulfide bonds Cys296-Cys389 and Cys368-Cys381 the C-terminal part of the tail is pulled back to a domain around position 296. This condensed negatively charged domain is ready for formation of salt-bridges with similar condensed opposite charged amino acids structures on the S1 RBD of SARS-CoV-2. This finding is fascinating and significant for a different reason to the others. It is not about Spike manipulation itself: in the next section we will explain that and how we believe that these capabilities were developed between 2008 - 2015. This finding points to something else: a trial to demonstrate a newly discovered attachment/co-receptor by field testing and verification. The context was the 2018 Swine Acute Diarrhoea Syndrome (SADS) outbreak in Guangzhou province.[10] Assuming that the Wuhan Institute of Virology team had discovered the functionalities of CLEC4M/DC-SIGN/CD209 receptors in the new SADS-CoV isolate and the fact that it could bind to positive charge (Ref: https://www.uniprot.org/uniprot/Q9NNX6 (CD209) and https://www.uniprot.org/uniprot/Q9H2X3) and that they wanted to do a field test of the described functionalities, the best conditions for doing so would be in connection with an ongoing viral infection. If this SADS originally did not have a ACE2 receptor binding motif (RBM), then a binding capacity verification of these attachment receptors could be done straightforwardly. But if SADS did have an ACE2 RBM, then it would be necessary to remove or disable the RBM of the Spike protein on this CoV isolate and execute the experiment in piglets including the formal Cox postulate verification of infection as described in the 2018 paper.
We postulate that there are 2 charged domains on SADS that are likely to contribute to attachment receptor binding located in domains 330-360 and 540-560 respectively. Recollect that we have identified a similar highly charged structure on SARS-CoV-2 within the edge of the RBD domain (526-560) with pI=10.03 which is brought right into the core of the RBD (to approximately position 400) by Cys-Cys bridging of the domain (538-590). This domain can contribute binding similar to that which can be observed for SADS. This new Cys-Cys property inserted into the SARS-CoV-2 Spike does not exist in SARS-CoV and hence could not provide such charge enhancement onto the RBD and co-receptor binding by natural evolution.
Taken all together, we suggest that our research findings on the general mode of action for infectivity of SARS-CoV-2 and the further puzzling features just mentioned, justify the question of the historical aetiology of these manipulations.
We did not need to address this issue diachronically for the purposes of vaccine design. However, it is important for a soundly based understanding of the present and potential future epidemiology of the Covid-19 pandemic and for strategies for its management. Therefore to our earlier amino-acid level of biochemical analysis we now add here a forensic analysis of published research literature concerning SARS-CoV-2. We will extend this type of analysis to three other viruses in the companion article.
Since, regrettably, international access has not been allowed to the relevant laboratories or materials, since Chinese scientists who wished to share their knowledge have not been able to do so and indeed since it appears that preserved virus material and related information have been destroyed, we are compelled to apply deduction to the published scientific literature, informed by our own biochemical analyses. We refute pre-emptively objection that this methodology does not result in absolute proof by observing that to make such a statement is to misunderstand scientific logic. The longer the chain of causation of individual findings that is shown, especially converging from different disciplines, the greater the confidence in the whole. We posit that the evidence below attains a high level of confidence.
A sequence of four linked research papers is explained
A comprehensive review of the relevant literature shows that a substantial amount of directly relevant gain-of function research has been undertaken. Four studies are especially noteworthy. They are linked in two ways: scientifically, in that the third and fourth build upon the results of the first and second, and in continuity of the institution and personnel across all four. The Wuhan Institute of Virology is a key collaborator in all these projects and Dr Zheng-Li Shi is one of the Institute's most experienced virologists and bat specialists. She is a common thread through all the key research projects.
Shi Zhengli
1. In 2008, Dr Shi was in the team whose research was an enabling pre-cursor to the two linked gain-of-function projects which lead to SARS-CoV-2's exact functionalities, including functionalities discovered via SADS and potentially field-tested in the 2018 study as suggested above. The 2008 Ren W et al project successfully demonstrated technical capabilities to interchange RBD’s between bat SARS-like and human SARS viruses: “... a minimal insert region (amino acids 310 to 518) was found to be sufficient to convert the SL-CoV S from non-ACE2 binding to human ACE2 binding, indicating that the SL-CoV S is largely compatible with SARS-CoV S protein both in structure and in function. The significance of these findings in relation to virus origin, virus recombination, and host switching is discussed" (Ren et al, 2008). Dr Shi is next a lead author of the second paper in this sequence, (Hou et al, 2010) and a co-author and the senior Chinese author of the third, (Menachery et al, 2015). She is also a co-author of the fourth (Zhou P. et al, 2018)
2. In 2010 scientists from the 'Special Viruses' section of the Wuhan Institute of Virology were engaged in 'gain of function' experiments, jointly with international collaborators, to increase SARS-CoV infectiousness for humans. They used an HIV pseudo virus to express seven bat ACE2 receptors and compared their binding properties to human ACE2 receptors in order to pick the best for further optimizing a SARS-like coronavirus’s ability to bind to human cells. They also found that some bat ACE2 receptors are very close to human ACE2 receptors. This study provided a model system for testing the most infectious of SARS-CoV-like viruses which already had been selected in a vast survey of Chinese bat populations between 2005 – 2013.(Xu L et al, 2016). These viruses were potentially infectious to humans via the ACE2 receptor. Further new viruses were identified between 2012-2015 (Lin et al,2017).
We came to the conclusion that there was manipulation around this virus. […] To a part but I do not say the total […] of the coronavirus of the bat, someone added sequences, in particular of HIV, the virus of AIDS. […] It is not natural. It’s the work of professionals, of molecular biologists. […] A very meticulous work.
-- Luc Montagnier, French virologist who won the Nobel Prize for Physiology or Medicine in 2008 for his discovery of the human immunodeficiency virus (HIV), during an appearance on France’s CNews, April 17, 2020"To evaluate the potential genetic changes required for HKU4 to infect human cells, we reengineered HKU4 spike, aiming to build its capacity to mediate viral entry into human cells. To this end, we introduced two single mutations, S746R and N762A, into HKU4 spike. The S746R mutation was expected to restore the hPPC motif in HKU4 spike, whereas the N762A mutation likely disrupted the potential N-linked glycosylation site in the hECP motif in HKU4 spike.
…
We examined the capability of the mutant HKU4 spike to mediate viral entry into three types of human cells (Fig. 3A for HEK293T cells; data not shown for Huh-7 and MRC-5 cells), using a pseudovirus entry assay as previously described (14). In the absence of exogenous protease trypsin, HKU4 pseudoviruses bearing either the reengineered hPPC motif or the reengineered hECP motif were able to enter human cells, whereas HKU4 pseudoviruses bearing both of the reengineered human protease motifs entered human cells as efficiently as when activated by exogenous trypsin (Fig. 3A). In contrast, wild-type HKU4 pseudoviruses failed to enter human cells. Therefore, the reengineered hPPC and hECP motifs enabled HKU4 spike to be activated by human endogenous proteases and thereby allowed HKU4 pseudoviruses to bypass the need for exogenous proteases to enter human cells. These results reveal that HKU4 spike needs only two single mutations at the S1/S2 boundary to gain the full capacity to mediate viral entry into human cells."
By the way, how they did it might frighten those who aren’t familiar with modern biotechnology — because the authors inserted this coronavirus spike-like protein into inactivated HIV:
"Briefly, MERS-CoV-spike-pseudotyped retroviruses expressing a luciferase reporter gene were prepared by cotransfecting HEK293T cells with a plasmid carrying Env-defective, luciferase-expressing HIV-1 genome (pNL4–3.luc.R-E-) and a plasmid encoding MERS-CoV spike protein."
Perhaps this is what prompted Indian researchers to look for sequences similar to HIV in the CoV2 genome (but their preprint was quickly criticized for bad methodology and erroneous conclusions). In fact, experts use such pseudoviruses regularly, and in general, one should not be scared of retroviruses as a class — their subspecies lentiviruses have been used for gene therapy for many years.
-- Lab-Made? SARS-CoV-2 Genealogy Through the Lens of Gain-of-Function Research, by Yuri Deigin
3. In 2015 scientists from the 'Special Viruses' section of the Wuhan Institute of Virology were engaged in 'gain of function' experiments jointly with a majority team from the University of North Carolina Chapel Hill. Together, they manipulated bat viruses to create a mouse adapted chimeric virus SHC014-MA15 which binds to and can proliferate on human upper airway cells (2B4 Calu-3 -- a cell line contributed by Chapel Hill): ("group 2b viruses encoding the SHC014 spike in a wild type backbone can efficiently utilize multiple ACE2 receptor orthologs, replicate efficiently in primary human airway cells, and achieve in vitro titers equivalent to epidemic strains of SARS-CoV"). We suggest that it is a high priority in further investigations to ascertain precisely from Chapel Hill lab records the exact donor provenance of 2B4 Calu-3. The lead Wuhan scientist, who provided the CoV material, was Dr Zheng-Li Shi ("provided SHC014 spike sequences and plasmids"). We note that what is described here are, in fact, precisely SARS-CoV-2 properties. In vivo experiments at Chapel Hill replicated the chimeric virus in mouse lung which showed significant pathogenesis which was the opposite of what the team had expected ("the creation of chimeric viruses like SHC014-MA15 was not expected to increase pathogenicity"). Menachery et al reported that it may be hard to develop a vaccine against SHC014-MA15. We can see, therefore, that the 2015 experiment advanced the 2010 work by perfecting in animal trials a virus optimised to infect the human upper respiratory tract. The 2015 authors were well aware that the chimeric virus which they had created was very dangerous because they discussed this fact. Of the opportunity/costs of their research, they suggested that “while offering preparation against future emerging viruses, this approach must be considered in the context of the US government-mandated pause on Gain Of Function (GOF) studies” (which has since been lifted). They also speculated that "review panels may deem similar studies too risky to pursue as increased pathogenicity in mammalian models cannot be excluded." It is certainly the case that this experiment created a chimeric virus with very high infectivity potential targeted to the human upper respiratory tract. Yet a surprising observation is that the paper states that this research consortium has permission to continue this research. It appears that optimisation gain of function work on this chimeric virus did continue. We deduce from paper authorships that this was done in the Wuhan Institute of Virology.
4. In 2018, as discussed earlier, Dr Shi's close colleague Peng Zhou, with others, investigated a coronavirus outbreak associated with a fatal Swine Acute Diarrhoea Syndrome (SADS) in Guangdong Province. This paper relates that piglets had a tissue specific infection site located in the intestine and that verification of the Bat Covid nature of this new SADS as the disease-causing agent was confirmed. 25,000 piglets died. However, the really interesting part of this study reports that in order to identify the receptor(s) used by the SADS CoV, known coronavirus host cell receptors were investigated: Angiotensin Converting Enzyme 2 (ACE2), Amino Peptidase N (APN), and Di-Peptidyl Peptidase 4 (DPP4). None of these receptors worked. But indirectly in their paper, the authors revealed their ability to express and to test new receptors in the ways posited earlier. Recollect that the model to do this was proven and reported in the 2010 work. Thus it is plain that SADS is a CoV infection utilising new tissue-specific binding domains; but the authors provide no hint about which receptor the virus is using in piglets except that it is not any of the best known three. We have offered our deduction above. Pigs, of course, have immune systems very similar to humans.
Now recollect that Menachery V.D et al in 2015 had shown that their chimeric virus SHC014-MA15 could, against their prediction, very successfully infect primary human upper airway epithelial cells (HAE) from the cell-line 2B4 Calu-3. With this in mind, we next observed that in the Covid-19 pandemic, a well-reported symptom in the early phase of the infection is loss of taste, headache and a sore throat. We have discussed this issue in the QRBD article in detail. But to summarise: in 2015 in a research review (Workman et al, 2015) discussed bitter/sweet taste receptors and the role these receptors play in mediating airway immune functions. They concluded thus: "Over the past several years, taste receptors have emerged as key players in the regulation of innate immune defenses in the mammalian respiratory tract. Several cell types in the airway, including ciliated epithelial cells, solitary chemosensory cells, and bronchial smooth muscle cells, all display chemoresponsive properties that utilize taste receptors."
Therefore we hypothesise the reconstructed historical aetiology of the Spike as follows:
In 2008, Dr Zheng-Li Si and WIV colleagues successfully demonstrated technical capabilities to interchange RBD’s between bat SARS-like and human SARS viruses. Building upon this, the 2010 work (Hou et al, 2010) perfected the ability to express receptors on human cells. On these foundations, the central Gain of Function work that underpins the functionalities of SARS-CoV-2 took place, carrying the WIV spike and plasmid materials to bond successfully to a UNC Chapel Hill human epithelial cell-line. This work (Menachery et al) produced a highly infectious chimeric virus optimised to the human upper respiratory tract. In convergent support of this hypothesis, both Lu (Lu et al, 2020) and Jia (Jia et al, 2020) have now, in January and April 2020, shown that SARS-CoV-2 has a bat SARS-like backbone but is carrying an RBD from a human SARS and Zhan et al have, like us, noted unusual adaption to humans from the first isolate. In the 2015 Chapel Hill work it was only ACE2 receptors that were discussed. However, in 2018 Zhou P. et al demonstrated capabilities to clone other receptors like APN and DPP4 and to test and compare these against the (intestine) tissue specific SADS-CoV identified. Then, in the 2019-20 Covid-19 pandemic, profuse symptoms indicating compromise of the bitter/sweet receptors are reported. Taken all together, this implies that by employing insights gained after 2015, as just deduced, a further optimization of the 2015 chimeric virus for additional binding to receptors/co-receptors such as bitter/sweet specific upper airway epithelia receptors occurred. That would help to explain the otherwise puzzling high infectivity and pathology associated with SARS-CoV-2 and hence also help to explain the social epidemiology of its spread.
Conclusion
We have deduced the internal logic of published research which resulted in the exact functionalities of SARS-CoV-2, including the convergence of agreement from difference classes of source, the timings of the stages of the research and the development of documented capabilities by named institutions and individuals. These meet the criteria of means, timing, agent and place in this reconstructed historical aetiology to produce sufficient confidence in the account to reverse the burden of proof. Henceforth, those who would maintain that the Covid-19 pandemic arose from zoonotic transfer need to explain precisely why this more parsimonious account is wrong before asserting that their evidence is persuasive, most especially when, as we have indicated, we note puzzling errors in their use of evidence. In our companion article, in a similar forensic manner we will explore the primary evidence used to sustain the hypothesis of zoonotic transfer. In neither this article nor the next do we speculate about motive.
Oslo & London 1 July 2020
_______________
References:
Alan D. Workman, James N. Palmer, Nithin D. Adappa and Noam A. Cohen. "The Role of Bitter and Sweet Taste Receptors in Upper Airway Immunity " Curr Allergy Asthma Rep. 2015 December ; 15(12): 72. doi:10.1007/s11882-015-0571-8.
Andersen, K. G., Rambaut, A., Lipkin, W.I., Holms, E. C., Garry, R. F., 2020, "The proximal origin of SARS-CoV-2". Nat Med. https://doi.org/10.1038/s41591-020-0820-9
Callaway E, "The Race for Coronavirus Vaccines", Nature, Vol 580, 30 April 2020, pp 576-7 (https://www.nature.com/articles/d41586-020-01221-y)
Hou Y, Peng C, Yu M, Li Y, Han Z, Li F, Wang LF, Shi Z., 2010, "Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry", Arch Virol, 155:1563–1569, doi: 10.1007/s00705-010-0729-6Hou 2010
Thomas-Oliver Kleen, Alicia A Galdon, Andrew S Macdonald and Angus G Dalgleish, Mitigating dysfunctional immunity in COVID-19: Trained Immunity, BCG and ‘new old friends’, 2020. (paper submitted)
Lin Xu, Fuqiang Zhang, Weihong Yang, Tinglei Jiang, Guanjun Lu, Biao He, Xingyu Li, Tingsong Hu, Gang Chen, Yun Feng, Yuzhen Zhang, Quanshui Fan, Jiang Feng, Hailin Zhang & Changchun Tu, 2016, "Detection and characterization of diverse alpha- and betacoronaviruses from bats in China", Virologica Sinica, 31 (1): 69–77, doi: 10.1007/s12250-016-3727-3
Marzi A, Gramberg T, Simmons G, Möller P, Rennekamp AJ, Krumbiegel M, Geier M, Eisemann J, Turza N, Saunier B, Steinkasserer A, Becker S, Bates P, Hofmann H, Pöhlmann S, (2004) "DC-SIGN and DC-SIGNR Interact with the Glycoprotein of Marburg Virus and the S Protein of Severe Acute Respiratory Syndrome Coronavirus", Journal of Virology, DOI: 10.1128/JVI.78.21.12090-12095.
Roujian Lu, Xiang Zhao, Juan Li, Peihua Niu, Bo Yang, Honglong Wu, Wenling Wang, Hao Song, Baoying Huang, Na Zhu, Yuhai Bi, Xuejun Ma, Faxian Zhan, Liang Wang, Tao Hu, Hong Zhou, Zhenhong Hu, Weimin Zhou, Li Zhao, Jing Chen, Yao Meng, Ji Wang, Yang Lin, Jianying Yuan, Zhihao Xie, Jinmin Ma, William J Liu, Dayan Wang, Wenbo Xu, Edward C Holmes, George F Gao, Guizhen Wu, Weijun Chen, Weifeng Shi, Wenjie Tan. “Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding”. Lancet 2020; 395: 565–74. https://doi.org/10.1016/ S0140-6736(20)30251-8
Sørensen, B., Susrud, A., & Dalgleish, A. (2020). "Biovacc-19: A Candidate Vaccine for Covid-19 (SARS-CoV-2) Developed from Analysis of its General Method of Action for Infectivity". QRB Discovery, 1-17. doi:10.1017/qrd.2020.8
Timothy Sheahan, Barry Rockx, Eric Donaldson, Amy Sims, Raymond Pickles, Davide Corti, Ralph Baric, 2008, “Mechanisms of Zoonotic Severe Acute Respiratory Syndrome Coronavirus Host Range Expansion in Human Airway Epithelium”. J Virol. https://doi.org/10.1128/JVI.02041-07
Vineet D Menachery, Boyd L Yount Jr, Kari Debbink, Sudhakar Agnihothram, Lisa E Gralinski, Jessica A Plante, Rachel L Graham, Trevor Scobey, Xing-Yi Ge, Eric F Donaldson, Scott H Randell, Antonio Lanzavecchia, Wayne A Marasco, Zhengli-Li Shi & Ralph S Baric , 2015," A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence", Nature Medicine, 21(12): 1508–1513. doi:10.1038/nm.3985
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D, (2020) “Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein”, Cell, Volume 181, Issue 2, 16 April, Pages 281-292.e6, DOI: https://doi.org/10.1016/j.cell.2020.02.058
Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. "Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus". J Virol 94: e00127-20. https://doi.org/10.1128/JVI.00127-20.
Wuze Ren, Xiuxia Qu, Wendong Li, Zhenggang Han, Meng Yu, Peng Zhou, Shu-Yi Zhang, Lin-Fa Wang, Hongkui Deng and Zhengli Shi, 2008, “Difference in Receptor Usage between Severe Acute Respiratory Syndrome (SARS) Coronavirus and SARS-Like Coronavirus of Bat Origin”. J Virol. https://doi.org/10.1128/JVI.01085-07
Xian-Dan Lin, Wen Wanga, Zong-Yu Hao, Zhao-Xiao Wang, Wen-Ping Guo, Xiao-Qing Guan, Miao-Ruo Wang, Hong-Wei Wang, Run-Hong Zhou, Ming-Hui Li, Guang-Peng Tang, Jun Wu, Edward C. Holmes, Yong-Zhen Zhang, 2017, "Extensive diversity of coronaviruses in bats from China", Virology 507, 1–10, https://doi.org/10.1016/j.virol.2017.03.019
Yong Jia, Gangxu Shen, Yujuan Zhang, Keng-Shiang Huang, Hsing-Ying Ho, Wei-Shio Hor, Chih-Hui Yang, Chengdao Li, Wei-Lung Wang. “Analysis of the mutation dynamics of SARS-CoV-2 reveals the spread history and emergence of RBD mutant with lower ACE2 binding affinity”. bioRxiv April 11,2020. https://doi.org/10.1101/2020.04.09.034942
Zhan S.H. et.al. (2020) "SARS-CoV-2 is well adapted for humans. What does this mean for re-emergence"- bioRxiv
Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XLL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG & Ma JY, (2018) "Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin", Nature 556, 255–258, https://doi.org/10.1038/s41586-018-0010-9
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF & Shi ZL, (2020) "A pneumonia outbreak associated with a new coronavirus of probable bat origin", Nature 579, 270–273, https://doi.org/10.1038/s41586-020-2012-7
- admin
- Site Admin
- Posts: 36135
- Joined: Thu Aug 01, 2013 5:21 am
Re: U.S. government gave $3.7 million grant to Wuhan lab at
Part 1 of 2
Why Do Exceptionally Dangerous Gain-of-Function Experiments in Influenza?
by Marc Lipsitch
Departments of Epidemiology and Immunology and Infectious Diseases, Center for Communicable Disease Dynamics, Harvard TH Chan School of Public Health, Boston, MA USA
Marc Lipsitch, Email: ude.dravrah.hpsh@ctispilm.
August 28, 2018
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
Abstract
This chapter makes the case against performing exceptionally dangerous gain-of-function experiments that are designed to create potentially pandemic and novel strains of influenza, for example, by enhancing the airborne transmissibility in mammals of highly virulent avian influenza strains. This is a question of intense debate over the last 5 years, though the history of such experiments goes back at least to the synthesis of viable influenza A H1N1 (1918) based on material preserved from the 1918 pandemic. This chapter makes the case that experiments to create potential pandemic pathogens (PPPs) are nearly unique in that they present biosafety risks that extend well beyond the experimenter or laboratory performing them; an accidental release could, as the name suggests, lead to global spread of a virulent virus, a biosafety incident on a scale never before seen. In such cases, biosafety considerations should be uppermost in the consideration of alternative approaches to experimental objectives and design, rather than being settled after the fact, as is appropriately done for most research involving pathogens. The extensive recent discussion of the magnitude of risks from such experiments is briefly reviewed. The chapter argues that, while there are indisputably certain questions that can be answered only by gain-of-function experiments in highly pathogenic strains, these questions are narrow and unlikely to meaningfully advance public health goals such as vaccine production and pandemic prediction. Alternative approaches to experimental influenza virology and characterization of existing strains are in general completely safe, higher throughput, more generalizable, and less costly than creation of PPP [Potential Pandemic Pathogens] in the laboratory and can thereby better inform public health. Indeed, virtually every finding of recent PPP [Potential Pandemic Pathogens] experiments that has been cited for its public health value was predated by similar findings using safe methodologies. The chapter concludes that the unique scientific and public health value of PPP [Potential Pandemic Pathogens] experiments is inadequate to justify the unique risks they entail and that researchers would be well-advised to turn their talents to other methodologies that will be safe and more rewarding scientifically.
Key words Influenza, Gain-of-function, Potential pandemic pathogen, Ferret, Evolution, Passage, Virulence, Transmissibility, Selection
The title of this chapter is not a rhetorical question. It is a serious question about what kinds of experiments influenza virologists should undertake. It is a privilege to be asked to address it in a laboratory manual that is primarily devoted to compiling protocols from leading virologists on how to perform experiments, once the scientific objective is selected.
This chapter expresses a view contrary to the view expressed by many leading influenza virologists. A few have argued that in the right hands, there are no exceptionally dangerous gain-of-function experiments in influenza. They argue that biosafety and biosecurity can be assured for such experiments when they are performed in the right laboratories, to a degree that the danger is not exceptional [1–3]. Others have argued that certain experiments likely to produce potential pandemic pathogens (PPPs)—defined as infectious agents capable of high virulence and efficient transmission between humans—are exceptionally valuable to basic science or to public health and are therefore worth whatever risk they may entail [1, 4, 5].
This chapter will make the case that, in most instances, both views are mistaken. Because this book is about virology methods, rather than about biosafety and biosecurity, the chapter will attend briefly to the risk side of the equation and will focus mainly on the question of scientific and public health value of these experiments. The argument will acknowledge that there are certain scientific questions that can be answered only by gain-of-function experiments and among these a subset that can be answered only by gain-of-function experiments that are expected to create PPPs [1, 4]. However it will attempt to convince the reader that the additional scientific value of this so-called gain-of-function research of concern (GoFRoC, in the elegant parlance of the US government) is relatively modest compared to what can be learned from GoF experiments that do not create PPPs, combined with other approaches to experimental and observational influenza studies [6, 7]. Last, it will make the claim that the value of this small class of GoFRoC experiments for public health is not, as has been claimed, exceptionally high but indeed is incremental at best and will not, in the foreseeable future, substantially improve the evidence base for public health decision-making [6].
For the purposes of this chapter, I will use interchangeably the phrases “exceptionally dangerous gain-of-function experiments”; “gain-of-function research of concern (GoFRoC),” as defined by the US National Science Advisory Board on Biosecurity; and “research likely to create PPP” as used in past writings by various authors. Specifically, these refer to experiments that are reasonably anticipated to create novel viral strains that combine high virulence for humans with high transmissibility in humans, especially when it is expected that the strain is likely to be unaffected by existing immunity in the general population due to antigenic novelty. Ferret passage of modified influenza A H5N1 viruses to select for droplet transmissibility [8, 9] is a paradigm case of such research. More generally, such studies (Fig. 1) typically start with a particular virus that does not have the phenotype of interest: to date, this has usually been droplet transmissibility between individuals of some mammalian species. Manipulations are performed to introduce changes that may confer such transmissibility. Such manipulations may involve (1) introducing (e.g., via reverse genetics) mutations expected to contribute to the phenotype [8], (2) providing novel genetic material by coinfecting cells or animals with another strain and permitting reassortment [10–12], or (3) simply waiting for spontaneous mutations to occur [8, 9, 13]. After performing one or more of these manipulations, the next step is to exert selection for the phenotype (say by placing an infected animal near but physically separated from an uninfected one and looking for viruses in the second animal that have moved via droplets from the first). The processes of introducing variation and selecting for the phenotype may be repeated one or more times and usually stop when the phenotype is identified. Different variations of these procedures have been used by several laboratories to enhance transmissibility of influenza A in various animal models [8–12].
Fig. 1 Schematic of the design of gain-of-function experiments involving selection for transmissibility in a mammalian host, starting from an avian-adapted virus obtained from an avian host or human zoonotic case
1. A Brief History of the Debate
Perhaps the first experimental effort to create a PPP in the laboratory was in fact by a somewhat different means: recreation of a strain of influenza H1N1 from 1918 by Tumpey and colleagues based on synthesizing nucleic acid sequences obtained from partially preserved viral RNA in frozen corpses from 1918 and then creating infectious virus by reverse genetics [14]. The question of whether it was wise to construct a virus that was historically associated with the worst pandemic in modern history and somewhat different from any virus currently circulating was raised, but the debate seems to have been internal to the US Department of Health and Human Services (HHS), particularly the National Institutes of Health (NIH), and it was judged that the work should proceed [14].
In 2004, as human cases of influenza H5N1 spilled over from avian reservoir hosts, a debate began about the desirability of experiments to test whether such viruses could reassort with human seasonal influenza viruses. Some argued that such experiments were essential and safe, while others argued for a global review of such studies, but apart from a news article in Science, there was little further public discussion [15].
Experiments to assess the potential for reassortment between subtypes of influenza enzootic in birds, such as H9N2 [10] and H5N1 [16, 17], continued with little attention from outside the field. Then, at a conference in Malta in September 2011, Prof. Ron Fouchier presented data from experiments in which his laboratory had modified a human isolate of H5N1 avian-origin influenza to acquire some mutations expected to adapt it to human-to-human transmission and then introduced the resulting virus into ferrets. Through serial passage in ferrets they had obtained a virus capable of small-droplet transmission from one infected ferret to another [3, 8]. Given the close resemblance of ferrets to humans in many aspects of influenza biology and pathology, this was seen by many (though not all [18]) as a proxy for human-to-human small-droplet transmission. Soon after, the laboratory of Prof. Yoshihiro Kawaoka reported a related set of experiments, this time using a virus created by reverse genetics from a human H1N1 virus and the hemagglutinin gene of a zoonotic H5N1 isolate [9]. A major controversy erupted over whether these experiments should be published. The controversy focused on the biosecurity issue of whether knowledge of the mutations required to render H5N1 influenza transmissible in ferrets (and possibly humans) would abet potential bioterrorists by providing them a “recipe” for a potential pandemic pathogen [19]. Despite strong objections from some, the US government, which had funded both sets of studies, recommended that they be published in full, as they soon were [8, 9]. Following this outcry, influenza virologists briefly announced [20, 21], then lifted [22, 23], a self-imposed moratorium on such experiments.
Concomitant with the publication of the two controversial studies, some colleagues raised concerns about a different risk: that of a laboratory accident that could lead to the release of a pathogen that, by design, combined high virulence and antigenic novelty (characteristic of the starting H5N1 strain) with high human-to-human transmissibility (a property selected for by proxy in the ferret experiments) [24–26]. These early critiques also questioned whether the scientific and public health value of such research justified the risks involved. Klotz and Sylvester coined the term “potential pandemic pathogen (PPP)” for such viruses [25]. The first major discussion meeting on this topic, to my knowledge, was held at the Royal Society of London in 2012 (https://royalsociety.org/science-events ... 2/viruses/), with mainly UK and North American speakers.
Following the revelations of a number of laboratory mishaps involving mishandling and in some cases possible human exposures to potentially lethal (but not highly transmissible) pathogens at high-containment federal laboratories in the United States, criticism of experiments that created PPP began to intensify. A meeting that I co-organized with colleagues in July 2014 led to the creation of the CambridgeWorking Group, which issued a statement calling for such work to be “curtailed” pending a formal risk-benefit assessment (http://www.cambridgeworkinggroup.org/). In response, a countervailing group called Scientists for Science (http://www.scientistsforscience.org/) formed in opposition to this call. Each group garnered the support of prominent scientists and others. In October 2014, citing the laboratory mishaps and the exceptional hazard that release of a potential pandemic pathogen might cause, the US White House Office of Science and Technology Policy (OSTP) and the National Institutes of Health announced a “pause” in federal funding of certain categories of research and a deliberative process that would culminate in a formal risk-benefit assessment, spearheaded by the National Science Advisory Board on Biosecurity (NSABB) (https://obamawhitehouse. archives.gov/blog/2014/10/17/doing-diligence-assess-risks-and-benefits-life-sciences-gain-function-research). This pause initially affected about two dozen existing grants from the NIH concerning influenza and coronaviruses [27], but it was soon narrowed to cover only about half that number (http://www. sciencemag.org/news/2014/12/moratorium-risky-experimentslifted-mers-mouse-studies), out of an NIH portfolio of about 250 active research grants at the time on these two groups of viruses.
Following the funding pause, two major discussion meetings were held by the US National Academy of Sciences [28, 29], multiple meetings were held by the NSABB (at the time of this writing, no link to the records of these meetings was available from https://osp.od.nih.gov/biotechnology/na ... ity-nsabb/), and a risk-benefit assessment commissioned by the NSABB was completed by a private consulting firm, Gryphon Scientific (http://www.gryphonscientific.com/wp-con ... Report.pdf [http://gryphonsci.wpengine.com/wp-content/uploads/2018/12/Risk-and-Benefit-Analysis-of-Gain-of-Function-Research-Final-Report-1.pdf]). Many scientists, a few ethicists, and others debated the scientific and public health rationale for PPP experiments, the risks they posed, and the ethics of doing research that poses potentially major risks to persons not involved in the studies and even unaware of them. This process raised awareness of many issues that had not been previously highlighted, notably the lack of a framework for evaluating risks of research to persons who are not research participants [30, 31], the very poor availability of data on biosafety in biological laboratories in the United States and elsewhere [32], and the consequent uncertainty in risk-benefit calculations.
In January 2017, OSTP announced that policy guidance for gain-of-function work was being released (https://www.phe.gov/s3/dualuse/Pages/Ga ... ction.aspx), and at the time of writing, there has been no final resolution of the issue.
This account has been focused on the US debate, with which I was most directly involved. A parallel process played out in Europe, slightly faster than in the United States, and with little net change in the conditions for such research to proceed. China, also an important player in PPP research, did not to my knowledge have a formal debate on the wisdom of such work, but this may reflect inadequate coverage in the Western press.
2. Making Choices About What Science to Do and How to Do It
The majority of this book is intended to facilitate the technical aspects of cutting-edge influenza virology by providing protocols and techniques for performing such experiments in the laboratory. This chapter, by contrast, attempts to address the (conceptually prior) question of which kinds of experiments one should undertake. Throughout a career in experimental biology, one makes choices about which experiments to perform—and which not to perform—on many different time scales. The choice of which field of biology to enter may have consequences for decades or an entire career. The choice of what project to select (from among those available in graduate school) or apply for grant support for (often at later stages of a career) will have consequences for many years; and the choices of which experimental approaches to take may govern one’s activities for an afternoon, a week, or several months or years depending on the complexity of the experiments. Over long time horizons, we make these decisions based on a combination of scientific curiosity, the potential for novelty, the importance of the problem for advancing science or achieving a practical (e.g., public health) goal, and more mundane but essential constraints such as the availability of technology, reagents, facilities, funding, mentors, and collaborators, to name a few. On the shorter time scale of hours, days, and months, the choice of detailed experimental approach is motivated by questions about which approach is:
• Most likely to provide interpretable data to answer the question, unaffected by uncontrolled variation
• Most likely to provide an answer fast
• Likely to be least expensive (or fit within a budget constraint)
• Most elegant scientifically
These are questions of judgment on which reasonable scientists will disagree because they have different tastes, skills, and resources, but they are the kinds of questions a funder or a principal investigator expects they will attempt to answer before proceeding.
Typically, biosafety has not been considered within this list of questions (except perhaps in the situation where time in a high-containment laboratory is a limiting resource). Rather, the scientific judgment is typically made first, and then appropriate biosafety conditions are identified to permit the research to be performed safely. In the specific case of US NIH grants, biosafety considerations are reviewed for a proposal only after the scientific merit is determined, and any concerns about biosafety are not supposed to affect the scientific score of a proposal. As a global research enterprise, this type of reasoning has worked relatively well: known fatalities associated with laboratory exposure to pathogens have been extremely rare [33], around one per year globally.
3. Unique Risks of GoF and GoFRoC Experiments
This procedure is not sufficient for those cases where the risk of a laboratory accident extends beyond those working in the laboratory or nearby—to a much larger, even global, population, who might be infected and greatly harmed by a pathogen that is both highly virulent to humans and readily transmissible [31]. As I will argue below, that category of research is a very limited category— roughly covered by the terms PPP creation or GoFRoC. The accident risk posed by performing an activity for a particular period of time is conventionally defined as the product of two factors: the probability that an accident will occur during that period, times the expected magnitude of harm that the accident causes. For the vast majority of pathogens studied in the laboratory, accidental infection of a laboratory worker or other person poses little risk to the broader population. This is because most pathogens we study have one or more of the following properties that limit the impact of an accidental infection:
• The pathogen is not transmissible from person to person (e.g., anthrax) or is somewhat transmissible but would be readily contained in a developed-world setting (e.g., filoviruses).
• The pathogen is already widespread (and possibly usually carried asymptomatically), such that adding one more infected person to the pool of infected persons would have a negligible impact on the overall risk of transmission (e.g., the bacterial species that constitute the normal flora of the gut and respiratory tract).
• Transmission of the pathogen is limited by widespread vaccine coverage (e.g., measles virus, Bordetella pertussis, and other targets of routine vaccination).
• Transmission of the pathogen is locally limited by lack of a suitable vector (e.g., flaviviruses in sufficiently cool climates).
Potential pandemic pathogens are intrinsically those which meet none of the above conditions: there is widespread susceptibility, high virulence for humans, little or no current human infection, and a route of transmission that is likely to be efficient in the population where the research is performed. In this case, the potential impact of an accidental infection is manyfold larger than for most pathogens under study. A probability of laboratory accident that may be acceptable when the impact is one or two human cases may not be acceptable when the potential impact is thousands, millions, or more [31].
There has been much debate about the exact magnitude of risk from GoFRoC experiments. Rehearsing those arguments is not the purpose of this chapter. A few points, however, are important:
1. A published estimate by the author of this chapter [34] using the experience of laboratory-acquired infections in BSL3 laboratories in the United States suggested that a single year of work in a single laboratory on a novel influenza virus readily transmissible between humans carries between a 1/1000 and a 1/10,000 chance of accidentally sparking a pandemic by accidental release. A reply by Prof. Ron Fouchier [2] placed the probability more than one million times lower. This is not the place to rehash the arguments over these probabilities. The essential point is that either estimate is a small probability; both are dependent on assumptions because data are limited; but when multiplied by the potential magnitude of a pandemic, the higher estimate (or even an estimate between the two) suggests a clearly unacceptable risk. While we believe that our estimate is if anything too low rather than too high, it is notable that the expected risk (probability of an accident leading to a pandemic times expected fatalities in such a pandemic) would be unacceptable to most people even if it were 1000-fold lower than we have estimated [34].
2. While excellent biosafety conditions in the laboratories performing GoFRoC are certainly important, it is not a panacea for guaranteeing safety. Of the major mishaps at US government labs in recent years, nearly all involved removing the infectious agent from the high-containment lab where it was under study to another, lower-containment lab because it was thought to be inert [35–37] (https://www.defense.gov/News/Special-Re ... ry-Review/). Hightech containment cannot prevent the deliberate removal of supposedly safe material from a laboratory, and so human error remains a source of potential missteps, regardless of the quality of the laboratory facilities. Among other consequences, this consideration means that measures to protect personnel in laboratories conducting GoFRoC, such as vaccination, prophylaxis with antiviral drugs, and enhanced surveillance for illness, cannot address the problem of exposure outside the “home” laboratory [38].
In summary, the record of laboratory accidents and accidental infections in the most secure and highly scrutinized government labs shows that such accidents are inevitable. In the vast majority of cases, the consequences, while potentially devastating for one or a few exposed persons, will be limited in scope. The unique risk posed by GoFRoC or PPP creation is that such accidents will lead not only to individual infections but to ongoing transmission and, in the worst case, extensive global spread of the engineered pathogen. Such risks are different in kind from ordinary biosafety risks and should not be seen as simply an issue to be addressed after deciding whether to do the experiment. Rather, the existence of such risks should be counted, like great expense in time or money, as an important factor arguing against undertaking the experiment in the first place. The unique benefits of doing GoFRoC should justify the unique risks such experiments create, and both comparisons should be made against devoting the same resources to alternative approaches that do not raise population-level biosafety concerns.
4. What Questions Can GoF and GoFRoC Experiments Answer Uniquely?
This leads directly to the question of what questions can be uniquely answered by experiments involving the creation of PPP. Here it is useful to distinguish the kinds of knowledge that can be obtained only by gain-of-function experiments and then the smaller set of knowledge that can be obtained only by GoFRoC, that is, by GOF experiments that lead to the creation of PPP.
Some scientific questions can be answered only by GoF experiments, as has been argued before [4]. However, these are limited to questions of the form: Starting from a particular virus genotype, is a particular set of genetic changes (for influenza, these may be reassortments or mutations) sufficient to create a particular phenotype that was not present in the starting genotype? This question can be answered only by a GoF experiment because if one does not create the phenotype, one cannot measure it. Loss-of-function experiments cannot answer this question unless one already has a strain with the phenotype in question and the exact genotype of interest, which is not the case in most GoF experiments. Comparisons of naturally occurring strains cannot answer the question for the same reason—if the phenotype already existed in a naturally occurring isolate, then the GoF experiment would not in general be under consideration.
Note the narrow scope of the question that such an experiment can answer. It cannot answer any of the following types of question:
1. Starting from a particular viral sequence, is a particular set of genetic changes necessary to create a phenotype of interest? This question is unanswerable in principle, because one can never make all possible genetic changes on a single starting genotype and rule out all alternative sets of changes [39].
2. Starting from a particular viral sequence, is a particular set of genetic changes the smallest set of changes on that genetic background sufficient to produce the phenotype? This question is unanswerable in practice for any set larger than about three mutations, because it is not possible to test phenotypically every one of the ~1012 combinations of three or more mutations, at least for whole-animal phenotypes like transmission that must be evaluated statistically in a number of animals for each genetic sequence. Thus even if each member of the set of changes is required along with the others to get the phenotype, there may be another set of (the same number of or fewer) changes that has not been tried in the experiment [39].
3. Is the set of mutations that produces the phenotype in the laboratory likely to happen in nature? This question can’t be answered by GoF experiments in the laboratory because viruses in nature are subject to many, possibly competing selective pressures and nonselective forces (e.g., population bottlenecks) that might produce a different trajectory of sequence changes from those that appear in the lab. Moreover, only a vanishingly small proportion of the viruses in nature have the genotype that forms the starting point of the experiment.
4. Is the set of mutations that produces the phenotype in the laboratory given a particular starting sequence likely to produce the phenotype when introduced into a viral strain with a different starting sequence? This would be predictable only if epistatic interactions were rare in influenza, such that the effect of a set of genetic changes on phenotype was relatively independent of genetic background. However, epistasis is pervasive in influenza [40, 41], including for phenotypes of interest such as and hemagglutinin receptor binding specificity [42, 43]. Thus, for example, the very mutations identified in GoF experiments on H5N1 isolates as conferring human receptor specificity on the viral hemagglutinin (HA) did not do so when introduced into a slightly different genetic background [43].
5. Does the set of mutations that confers ferret-to-ferret droplet transmissibility when introduced into a particular starting sequence also confer human-to-human transmissibility? Barring an unethical deliberate infection of humans with the virus resulting from GoF experiments in ferrets, and barring an accidental infection of humans, which would be potentially catastrophic, we will never know whether that particular virus’s transmission potential in ferrets translates to humans. Ferret droplet transmissibility is a very good, but not perfect predictor of human transmissibility for influenza [44]. At least one prominent influenza virologist has argued that the strains produced in GoF experiments in ferrets probably are not readily transmissible in humans [18], and others have said the same thing in public forums. If taken seriously, this uncertainty undermines the scientific and public health value of the experiments, because it undermines our ability to use the presence of GoF-identified mutations and phenotypes to gauge the level of risk presented by naturally occurring viruses [45].
4.1 What Scientific Questions Are of Greatest Importance for Pandemic Prevention and Response?
Each of the above classes of questions—those answerable by GoF experiments (in some cases by GoFRoC experiments) and those not answerable by such experiments—has legitimate scientific interest. But improving our ability to prevent and/or respond to novel influenza pandemics is the major public health goal that lends practical importance to advancing our scientific knowledge of the determinants of transmissibility in particular. Most importantly, effective targeting of efforts to prevent or mitigate pandemic threats partly depends on developing accurate predictive capabilities to estimate the magnitude of the threat posed by particular influenza A strains that we are observing in nonhuman reservoirs [40, 45]. Knowing that strains isolated from wild or domesticated avian or domesticated swine populations have genetic sequences or phenotypic traits (e.g., receptor binding specificity) predictive of the ability to transmit effectively in humans can motivate targeted countermeasures against such strains, such as culling of infected animals or strain-specific vaccine development. These two activities in particular depend on prioritizing which strains are most threatening, because it is impractical to cull all influenza-infected animals, and it is currently impractical to make vaccines except in a strainspecific fashion. Such strain-dependent prevention and mitigation activities are part of an overall pandemic preparedness portfolio including strengthening human respiratory disease surveillance and analytic capacity, stockpiling of non-strain-specific countermeasures such as antiviral drugs, personal protective equipment and ventilators, and a range of other public health measures whose usefulness is relatively independent of the strain of influenza that causes zoonotic cases or initiates a pandemic. But for animal culling and vaccine development (absent universal vaccines), prioritizing pandemic threats based on sequence and viral traits is important [40, 45, 46].
Many different types of scientific approaches can contribute to such prioritization efforts [7]. Comparison of naturally occurring influenza viruses, either by genetic sequence or by phenotypic traits or both, can help to categorize those sequences and traits that are characteristics of strains that are avian-adapted and poorly able to transmit in humans, compared to those that are well-adapted to human transmission. Such comparisons have been made for decades, and a number of traits and genetic sequence features (such as specific variants at specific amino acid sites in specific viral proteins) have been associated with human adaptation. Among the most important traits correlated with human adaptation are HA specificity for sialic acids found in the human upper airway (alpha-2,6 linked), a relatively low pH of HA activation, and human adaptation of the nucleoprotein-polymerase complex [40].
As the importance of these and other traits has become apparent from natural comparisons, the sequence variations correlated with these properties have been established through biochemical and other assays of single gene products or minigenomes (in the case of the nucleoprotein-polymerase complex) [47]. Such sequence changes can then be tested for their causal role in the individual traits, measurable in vitro, again without employing infectious virus [48–50]. Gain-of-function studies can be performed without employing strains with pandemic potential, for example, by taking a natural strain, introducing a change that abrogates one of the traits of interest, and then selecting revertants (so-called gain-loss-of-function) [51] or by doing gain-of-function experiments similar to GoFRoC but employing attenuated strains [52], strains of lower virulence, and strains for which there is greater population-wide immunity. All of these are alternatives to GoFRoC, and each is capable of informing public health efforts to prioritize pandemic threats from naturally occurring strains.
Importantly, such prioritization efforts are and will remain intrinsically imperfect. There have only been four influenza pandemics since the turn of the twentieth century, so our evolutionary models of what confers human-to-human transmissibility are constrained by being based on essentially four independent data points. The limitations of predicting pandemic potential were evident in 2009, when the strain that emerged and began spreading in humans arguably failed to meet the existing notions of human adaptation on all three key human-adaptive traits as understood at the time: early pandemic isolates’ HA receptor specificity was mixed human and avian; their HA pH of activation was outside the range of human-adapted viruses known before; and they lacked the sequence variant (lysine at PB2 amino acid 627) that had been thought necessary in the past for human adaptation. While subsequent evolution “corrected” the first two of these limitations on human-to-human transmission, the criteria had to be updated to accommodate the fact that what seemed necessary for pandemic potential in 2008 was violated by the early pandemic strain in 2009 [40]. While this limitation is surely a rationale for continuing to expand our scientific knowledge in this area, it more fundamentally points out that with only four modern pandemics to test our models of what is needed for a pandemic strain, no amount of further research will be able to make a highly reliable predictive model for pandemic potential—we simply lack the data (actual pandemics) to validate such models and therefore can never rely fully on these models, no matter how many more experimental inputs we add. This consideration heightens the importance of other approaches to pandemic preparedness, which are not dependent on correct predictions of the strain that will cause a pandemic, in combination with efforts to improve our predictive power. Table 1 lists a number of experimental and computational approaches to increasing our understanding of influenza pandemic risk determinants (top) and increasing our preparedness for pandemics more generally (bottom).
Why Do Exceptionally Dangerous Gain-of-Function Experiments in Influenza?
by Marc Lipsitch
Departments of Epidemiology and Immunology and Infectious Diseases, Center for Communicable Disease Dynamics, Harvard TH Chan School of Public Health, Boston, MA USA
Marc Lipsitch, Email: ude.dravrah.hpsh@ctispilm.
August 28, 2018
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
YOU ARE REQUIRED TO READ THE COPYRIGHT NOTICE AT THIS LINK BEFORE YOU READ THE FOLLOWING WORK, THAT IS AVAILABLE SOLELY FOR PRIVATE STUDY, SCHOLARSHIP OR RESEARCH PURSUANT TO 17 U.S.C. SECTION 107 AND 108. IN THE EVENT THAT THE LIBRARY DETERMINES THAT UNLAWFUL COPYING OF THIS WORK HAS OCCURRED, THE LIBRARY HAS THE RIGHT TO BLOCK THE I.P. ADDRESS AT WHICH THE UNLAWFUL COPYING APPEARED TO HAVE OCCURRED. THANK YOU FOR RESPECTING THE RIGHTS OF COPYRIGHT OWNERS.
-- Ethical Alternatives to Experiments with Novel Potential Pandemic Pathogens, by Marc Lipsitch, Allison P. Galvani
-- Risk and Benefit Analysis of Gain of Function Research, Final Report, April 2016, by Gryphon Scientific
-- Review Committee Report: Inadvertent Shipment of Live Bacillus anthracis Spores by DoD, July 13, 2015
Abstract
This chapter makes the case against performing exceptionally dangerous gain-of-function experiments that are designed to create potentially pandemic and novel strains of influenza, for example, by enhancing the airborne transmissibility in mammals of highly virulent avian influenza strains. This is a question of intense debate over the last 5 years, though the history of such experiments goes back at least to the synthesis of viable influenza A H1N1 (1918) based on material preserved from the 1918 pandemic. This chapter makes the case that experiments to create potential pandemic pathogens (PPPs) are nearly unique in that they present biosafety risks that extend well beyond the experimenter or laboratory performing them; an accidental release could, as the name suggests, lead to global spread of a virulent virus, a biosafety incident on a scale never before seen. In such cases, biosafety considerations should be uppermost in the consideration of alternative approaches to experimental objectives and design, rather than being settled after the fact, as is appropriately done for most research involving pathogens. The extensive recent discussion of the magnitude of risks from such experiments is briefly reviewed. The chapter argues that, while there are indisputably certain questions that can be answered only by gain-of-function experiments in highly pathogenic strains, these questions are narrow and unlikely to meaningfully advance public health goals such as vaccine production and pandemic prediction. Alternative approaches to experimental influenza virology and characterization of existing strains are in general completely safe, higher throughput, more generalizable, and less costly than creation of PPP [Potential Pandemic Pathogens] in the laboratory and can thereby better inform public health. Indeed, virtually every finding of recent PPP [Potential Pandemic Pathogens] experiments that has been cited for its public health value was predated by similar findings using safe methodologies. The chapter concludes that the unique scientific and public health value of PPP [Potential Pandemic Pathogens] experiments is inadequate to justify the unique risks they entail and that researchers would be well-advised to turn their talents to other methodologies that will be safe and more rewarding scientifically.
Ralph Nader: Before we get to the coronavirus COVID-19 aspect of our discussion, let's start with your point about lack of public knowledge or debate about what researchers around the world are genetically engineering.
Andrew Kimbrell: Yeah, Ralph, I think this is really just an absolutely critical issue that's been lost -- the forest has been lost for the trees, if you will. We have talked on your show, and you've worked on this a lot, about the dangers of genetic engineering of plants, animals, or humans. But we haven't talked much about the danger of genetically engineering viruses. And I think far away from public debate, a small group of scientists over the last 10 or 11 years have used synthetic biology, synthetic virology, to be able to do something which is breathtaking. What they've done is they've taken the most dangerous viruses known to man -- these are H5N1, bird flu, Marburg, Ebola, SARS -- and instead of trying to find vaccines, or to make these viruses less lethal, they have actually spent millions and millions of our taxpayer dollars, tens of millions of our taxpayer dollars, trying to make these viruses more dangerous, by mixing and matching various parts of these viruses with other viruses, genetically engineering them, and then using animal experimentation, and human cell line experimentation, to make them more transmissible, to make them more lethal, to make them more infectious.
Ralph Nader: Scientifically, why would they want to do this?
Andrew Kimbrell: Well, I do have a master's degree in psychology, and I think I have to sort of rely on that to try and figure out why anybody would want to do this. One prominent virologist has called it the definition of insanity. I think there is a temptation, and this probably goes back, Ralph, to the creation of nuclear weapons, the early experimentations that we all saw with genetic engineering, putting human genes into pigs, and doing all sorts of crazy things. There's this problem with some of our scientists that just because you can do something, they think that you should do it. There really is no end. Marc Lipsitch from Harvard, and Thomas Inglesby, a prominent health security expert at Johns Hopkins, they've gone to great lengths to show there's no value, that we've actually gotten zero value from these experiments. The [scientists] say, "Hey, if we create these novel, brand new pandemic viruses, maybe nature will create them later, and we'll have some kind of intervention strategy for them." But that makes no sense, of course, because nature has a million different variations that we would never be able to predict. So the idea that we can somehow predict in the laboratory, and then spend tens of millions of dollars trying to find an intervention strategy when there could be millions of other combinations out there in nature, makes no sense. There is one unfortunate place where this kind of research could be useful, and that would be the creation of biological weapons....
Let me just give you an example. So there's something called H5N1 bird flu. Most people have heard of it. Just a few hundred people have been infected by it, but it has a 60% mortality. Whoever gets it, 60% of the people die. Compare that to, for example, what's happening with COVID-19; some people say it's 1%, 4%, we'll see. But imagine 60%. Well, two researchers, Ron Fouchier who's up at the University of Erasmus in Netherlands, and Yoshihiro Kawaoka who is a researcher at University of Wisconsin, they said, "You know what, this isn't very infective, this bird flu. What if we were able to create a version that is airborne? You could get like the common cold. Let's try that." And they did. They actually were able to create this virus. So if this virus escapes, right, 1.6 billion people could die, 60% of the world's population. Well, this caused a huge furor. In 2014, the Obama administration actually declared a moratorium on this gain of function--gain of threat. I don't like calling it gain of function because that's euphemistic; it's gain of threat research, great threat, creating novel pandemic viruses. They said, "This is just too dangerous."
-- Interview with Andrew Kimball on the Ralph Nader Radio Show, July 18, 2020
Key words Influenza, Gain-of-function, Potential pandemic pathogen, Ferret, Evolution, Passage, Virulence, Transmissibility, Selection
The title of this chapter is not a rhetorical question. It is a serious question about what kinds of experiments influenza virologists should undertake. It is a privilege to be asked to address it in a laboratory manual that is primarily devoted to compiling protocols from leading virologists on how to perform experiments, once the scientific objective is selected.
This chapter expresses a view contrary to the view expressed by many leading influenza virologists. A few have argued that in the right hands, there are no exceptionally dangerous gain-of-function experiments in influenza. They argue that biosafety and biosecurity can be assured for such experiments when they are performed in the right laboratories, to a degree that the danger is not exceptional [1–3]. Others have argued that certain experiments likely to produce potential pandemic pathogens (PPPs)—defined as infectious agents capable of high virulence and efficient transmission between humans—are exceptionally valuable to basic science or to public health and are therefore worth whatever risk they may entail [1, 4, 5].
This chapter will make the case that, in most instances, both views are mistaken. Because this book is about virology methods, rather than about biosafety and biosecurity, the chapter will attend briefly to the risk side of the equation and will focus mainly on the question of scientific and public health value of these experiments. The argument will acknowledge that there are certain scientific questions that can be answered only by gain-of-function experiments and among these a subset that can be answered only by gain-of-function experiments that are expected to create PPPs [1, 4]. However it will attempt to convince the reader that the additional scientific value of this so-called gain-of-function research of concern (GoFRoC, in the elegant parlance of the US government) is relatively modest compared to what can be learned from GoF experiments that do not create PPPs, combined with other approaches to experimental and observational influenza studies [6, 7]. Last, it will make the claim that the value of this small class of GoFRoC experiments for public health is not, as has been claimed, exceptionally high but indeed is incremental at best and will not, in the foreseeable future, substantially improve the evidence base for public health decision-making [6].
For the purposes of this chapter, I will use interchangeably the phrases “exceptionally dangerous gain-of-function experiments”; “gain-of-function research of concern (GoFRoC),” as defined by the US National Science Advisory Board on Biosecurity; and “research likely to create PPP” as used in past writings by various authors. Specifically, these refer to experiments that are reasonably anticipated to create novel viral strains that combine high virulence for humans with high transmissibility in humans, especially when it is expected that the strain is likely to be unaffected by existing immunity in the general population due to antigenic novelty. Ferret passage of modified influenza A H5N1 viruses to select for droplet transmissibility [8, 9] is a paradigm case of such research. More generally, such studies (Fig. 1) typically start with a particular virus that does not have the phenotype of interest: to date, this has usually been droplet transmissibility between individuals of some mammalian species. Manipulations are performed to introduce changes that may confer such transmissibility. Such manipulations may involve (1) introducing (e.g., via reverse genetics) mutations expected to contribute to the phenotype [8], (2) providing novel genetic material by coinfecting cells or animals with another strain and permitting reassortment [10–12], or (3) simply waiting for spontaneous mutations to occur [8, 9, 13]. After performing one or more of these manipulations, the next step is to exert selection for the phenotype (say by placing an infected animal near but physically separated from an uninfected one and looking for viruses in the second animal that have moved via droplets from the first). The processes of introducing variation and selecting for the phenotype may be repeated one or more times and usually stop when the phenotype is identified. Different variations of these procedures have been used by several laboratories to enhance transmissibility of influenza A in various animal models [8–12].

Fig. 1 Schematic of the design of gain-of-function experiments involving selection for transmissibility in a mammalian host, starting from an avian-adapted virus obtained from an avian host or human zoonotic case
1. A Brief History of the Debate
Perhaps the first experimental effort to create a PPP in the laboratory was in fact by a somewhat different means: recreation of a strain of influenza H1N1 from 1918 by Tumpey and colleagues based on synthesizing nucleic acid sequences obtained from partially preserved viral RNA in frozen corpses from 1918 and then creating infectious virus by reverse genetics [14]. The question of whether it was wise to construct a virus that was historically associated with the worst pandemic in modern history and somewhat different from any virus currently circulating was raised, but the debate seems to have been internal to the US Department of Health and Human Services (HHS), particularly the National Institutes of Health (NIH), and it was judged that the work should proceed [14].
In 2004, as human cases of influenza H5N1 spilled over from avian reservoir hosts, a debate began about the desirability of experiments to test whether such viruses could reassort with human seasonal influenza viruses. Some argued that such experiments were essential and safe, while others argued for a global review of such studies, but apart from a news article in Science, there was little further public discussion [15].
Experiments to assess the potential for reassortment between subtypes of influenza enzootic in birds, such as H9N2 [10] and H5N1 [16, 17], continued with little attention from outside the field. Then, at a conference in Malta in September 2011, Prof. Ron Fouchier presented data from experiments in which his laboratory had modified a human isolate of H5N1 avian-origin influenza to acquire some mutations expected to adapt it to human-to-human transmission and then introduced the resulting virus into ferrets. Through serial passage in ferrets they had obtained a virus capable of small-droplet transmission from one infected ferret to another [3, 8]. Given the close resemblance of ferrets to humans in many aspects of influenza biology and pathology, this was seen by many (though not all [18]) as a proxy for human-to-human small-droplet transmission. Soon after, the laboratory of Prof. Yoshihiro Kawaoka reported a related set of experiments, this time using a virus created by reverse genetics from a human H1N1 virus and the hemagglutinin gene of a zoonotic H5N1 isolate [9]. A major controversy erupted over whether these experiments should be published. The controversy focused on the biosecurity issue of whether knowledge of the mutations required to render H5N1 influenza transmissible in ferrets (and possibly humans) would abet potential bioterrorists by providing them a “recipe” for a potential pandemic pathogen [19]. Despite strong objections from some, the US government, which had funded both sets of studies, recommended that they be published in full, as they soon were [8, 9]. Following this outcry, influenza virologists briefly announced [20, 21], then lifted [22, 23], a self-imposed moratorium on such experiments.
Concomitant with the publication of the two controversial studies, some colleagues raised concerns about a different risk: that of a laboratory accident that could lead to the release of a pathogen that, by design, combined high virulence and antigenic novelty (characteristic of the starting H5N1 strain) with high human-to-human transmissibility (a property selected for by proxy in the ferret experiments) [24–26]. These early critiques also questioned whether the scientific and public health value of such research justified the risks involved. Klotz and Sylvester coined the term “potential pandemic pathogen (PPP)” for such viruses [25]. The first major discussion meeting on this topic, to my knowledge, was held at the Royal Society of London in 2012 (https://royalsociety.org/science-events ... 2/viruses/), with mainly UK and North American speakers.
Following the revelations of a number of laboratory mishaps involving mishandling and in some cases possible human exposures to potentially lethal (but not highly transmissible) pathogens at high-containment federal laboratories in the United States, criticism of experiments that created PPP began to intensify. A meeting that I co-organized with colleagues in July 2014 led to the creation of the CambridgeWorking Group, which issued a statement calling for such work to be “curtailed” pending a formal risk-benefit assessment (http://www.cambridgeworkinggroup.org/). In response, a countervailing group called Scientists for Science (http://www.scientistsforscience.org/) formed in opposition to this call. Each group garnered the support of prominent scientists and others. In October 2014, citing the laboratory mishaps and the exceptional hazard that release of a potential pandemic pathogen might cause, the US White House Office of Science and Technology Policy (OSTP) and the National Institutes of Health announced a “pause” in federal funding of certain categories of research and a deliberative process that would culminate in a formal risk-benefit assessment, spearheaded by the National Science Advisory Board on Biosecurity (NSABB) (https://obamawhitehouse. archives.gov/blog/2014/10/17/doing-diligence-assess-risks-and-benefits-life-sciences-gain-function-research). This pause initially affected about two dozen existing grants from the NIH concerning influenza and coronaviruses [27], but it was soon narrowed to cover only about half that number (http://www. sciencemag.org/news/2014/12/moratorium-risky-experimentslifted-mers-mouse-studies), out of an NIH portfolio of about 250 active research grants at the time on these two groups of viruses.
Following the funding pause, two major discussion meetings were held by the US National Academy of Sciences [28, 29], multiple meetings were held by the NSABB (at the time of this writing, no link to the records of these meetings was available from https://osp.od.nih.gov/biotechnology/na ... ity-nsabb/), and a risk-benefit assessment commissioned by the NSABB was completed by a private consulting firm, Gryphon Scientific (http://www.gryphonscientific.com/wp-con ... Report.pdf [http://gryphonsci.wpengine.com/wp-content/uploads/2018/12/Risk-and-Benefit-Analysis-of-Gain-of-Function-Research-Final-Report-1.pdf]). Many scientists, a few ethicists, and others debated the scientific and public health rationale for PPP experiments, the risks they posed, and the ethics of doing research that poses potentially major risks to persons not involved in the studies and even unaware of them. This process raised awareness of many issues that had not been previously highlighted, notably the lack of a framework for evaluating risks of research to persons who are not research participants [30, 31], the very poor availability of data on biosafety in biological laboratories in the United States and elsewhere [32], and the consequent uncertainty in risk-benefit calculations.
In January 2017, OSTP announced that policy guidance for gain-of-function work was being released (https://www.phe.gov/s3/dualuse/Pages/Ga ... ction.aspx), and at the time of writing, there has been no final resolution of the issue.
This account has been focused on the US debate, with which I was most directly involved. A parallel process played out in Europe, slightly faster than in the United States, and with little net change in the conditions for such research to proceed. China, also an important player in PPP research, did not to my knowledge have a formal debate on the wisdom of such work, but this may reflect inadequate coverage in the Western press.
2. Making Choices About What Science to Do and How to Do It
The majority of this book is intended to facilitate the technical aspects of cutting-edge influenza virology by providing protocols and techniques for performing such experiments in the laboratory. This chapter, by contrast, attempts to address the (conceptually prior) question of which kinds of experiments one should undertake. Throughout a career in experimental biology, one makes choices about which experiments to perform—and which not to perform—on many different time scales. The choice of which field of biology to enter may have consequences for decades or an entire career. The choice of what project to select (from among those available in graduate school) or apply for grant support for (often at later stages of a career) will have consequences for many years; and the choices of which experimental approaches to take may govern one’s activities for an afternoon, a week, or several months or years depending on the complexity of the experiments. Over long time horizons, we make these decisions based on a combination of scientific curiosity, the potential for novelty, the importance of the problem for advancing science or achieving a practical (e.g., public health) goal, and more mundane but essential constraints such as the availability of technology, reagents, facilities, funding, mentors, and collaborators, to name a few. On the shorter time scale of hours, days, and months, the choice of detailed experimental approach is motivated by questions about which approach is:
• Most likely to provide interpretable data to answer the question, unaffected by uncontrolled variation
• Most likely to provide an answer fast
• Likely to be least expensive (or fit within a budget constraint)
• Most elegant scientifically
These are questions of judgment on which reasonable scientists will disagree because they have different tastes, skills, and resources, but they are the kinds of questions a funder or a principal investigator expects they will attempt to answer before proceeding.
Typically, biosafety has not been considered within this list of questions (except perhaps in the situation where time in a high-containment laboratory is a limiting resource). Rather, the scientific judgment is typically made first, and then appropriate biosafety conditions are identified to permit the research to be performed safely. In the specific case of US NIH grants, biosafety considerations are reviewed for a proposal only after the scientific merit is determined, and any concerns about biosafety are not supposed to affect the scientific score of a proposal. As a global research enterprise, this type of reasoning has worked relatively well: known fatalities associated with laboratory exposure to pathogens have been extremely rare [33], around one per year globally.
3. Unique Risks of GoF and GoFRoC Experiments
This procedure is not sufficient for those cases where the risk of a laboratory accident extends beyond those working in the laboratory or nearby—to a much larger, even global, population, who might be infected and greatly harmed by a pathogen that is both highly virulent to humans and readily transmissible [31]. As I will argue below, that category of research is a very limited category— roughly covered by the terms PPP creation or GoFRoC. The accident risk posed by performing an activity for a particular period of time is conventionally defined as the product of two factors: the probability that an accident will occur during that period, times the expected magnitude of harm that the accident causes. For the vast majority of pathogens studied in the laboratory, accidental infection of a laboratory worker or other person poses little risk to the broader population. This is because most pathogens we study have one or more of the following properties that limit the impact of an accidental infection:
• The pathogen is not transmissible from person to person (e.g., anthrax) or is somewhat transmissible but would be readily contained in a developed-world setting (e.g., filoviruses).
• The pathogen is already widespread (and possibly usually carried asymptomatically), such that adding one more infected person to the pool of infected persons would have a negligible impact on the overall risk of transmission (e.g., the bacterial species that constitute the normal flora of the gut and respiratory tract).
• Transmission of the pathogen is limited by widespread vaccine coverage (e.g., measles virus, Bordetella pertussis, and other targets of routine vaccination).
• Transmission of the pathogen is locally limited by lack of a suitable vector (e.g., flaviviruses in sufficiently cool climates).
Potential pandemic pathogens are intrinsically those which meet none of the above conditions: there is widespread susceptibility, high virulence for humans, little or no current human infection, and a route of transmission that is likely to be efficient in the population where the research is performed. In this case, the potential impact of an accidental infection is manyfold larger than for most pathogens under study. A probability of laboratory accident that may be acceptable when the impact is one or two human cases may not be acceptable when the potential impact is thousands, millions, or more [31].
Let me just give you an example. So there's something called H5N1 bird flu. Most people have heard of it. Just a few hundred people have been infected by it, but it has a 60% mortality. Whoever gets it, 60% of the people die. Compare that to, for example, what's happening with COVID-19; some people say it's 1%, 4%, we'll see. But imagine 60%. Well, two researchers, Ron Fouchier who's up at the University of Erasmus in Netherlands, and Yoshihiro Kawaoka who is a researcher at University of Wisconsin, they said, "You know what, this isn't very infective, this bird flu. What if we were able to create a version that is airborne? You could get like the common cold. Let's try that." And they did. They actually were able to create this virus. So if this virus escapes, right, 1.6 billion people could die, 60% of the world's population.
-- Interview with Andrew Kimball on the Ralph Nader Radio Show, July 18, 2020
There has been much debate about the exact magnitude of risk from GoFRoC experiments. Rehearsing those arguments is not the purpose of this chapter. A few points, however, are important:
1. A published estimate by the author of this chapter [34] using the experience of laboratory-acquired infections in BSL3 laboratories in the United States suggested that a single year of work in a single laboratory on a novel influenza virus readily transmissible between humans carries between a 1/1000 and a 1/10,000 chance of accidentally sparking a pandemic by accidental release. A reply by Prof. Ron Fouchier [2] placed the probability more than one million times lower. This is not the place to rehash the arguments over these probabilities. The essential point is that either estimate is a small probability; both are dependent on assumptions because data are limited; but when multiplied by the potential magnitude of a pandemic, the higher estimate (or even an estimate between the two) suggests a clearly unacceptable risk. While we believe that our estimate is if anything too low rather than too high, it is notable that the expected risk (probability of an accident leading to a pandemic times expected fatalities in such a pandemic) would be unacceptable to most people even if it were 1000-fold lower than we have estimated [34].
2. While excellent biosafety conditions in the laboratories performing GoFRoC are certainly important, it is not a panacea for guaranteeing safety. Of the major mishaps at US government labs in recent years, nearly all involved removing the infectious agent from the high-containment lab where it was under study to another, lower-containment lab because it was thought to be inert [35–37] (https://www.defense.gov/News/Special-Re ... ry-Review/). Hightech containment cannot prevent the deliberate removal of supposedly safe material from a laboratory, and so human error remains a source of potential missteps, regardless of the quality of the laboratory facilities. Among other consequences, this consideration means that measures to protect personnel in laboratories conducting GoFRoC, such as vaccination, prophylaxis with antiviral drugs, and enhanced surveillance for illness, cannot address the problem of exposure outside the “home” laboratory [38].
In summary, the record of laboratory accidents and accidental infections in the most secure and highly scrutinized government labs shows that such accidents are inevitable. In the vast majority of cases, the consequences, while potentially devastating for one or a few exposed persons, will be limited in scope. The unique risk posed by GoFRoC or PPP creation is that such accidents will lead not only to individual infections but to ongoing transmission and, in the worst case, extensive global spread of the engineered pathogen. Such risks are different in kind from ordinary biosafety risks and should not be seen as simply an issue to be addressed after deciding whether to do the experiment. Rather, the existence of such risks should be counted, like great expense in time or money, as an important factor arguing against undertaking the experiment in the first place. The unique benefits of doing GoFRoC should justify the unique risks such experiments create, and both comparisons should be made against devoting the same resources to alternative approaches that do not raise population-level biosafety concerns.
4. What Questions Can GoF and GoFRoC Experiments Answer Uniquely?
This leads directly to the question of what questions can be uniquely answered by experiments involving the creation of PPP. Here it is useful to distinguish the kinds of knowledge that can be obtained only by gain-of-function experiments and then the smaller set of knowledge that can be obtained only by GoFRoC, that is, by GOF experiments that lead to the creation of PPP.
Some scientific questions can be answered only by GoF experiments, as has been argued before [4]. However, these are limited to questions of the form: Starting from a particular virus genotype, is a particular set of genetic changes (for influenza, these may be reassortments or mutations) sufficient to create a particular phenotype that was not present in the starting genotype? This question can be answered only by a GoF experiment because if one does not create the phenotype, one cannot measure it. Loss-of-function experiments cannot answer this question unless one already has a strain with the phenotype in question and the exact genotype of interest, which is not the case in most GoF experiments. Comparisons of naturally occurring strains cannot answer the question for the same reason—if the phenotype already existed in a naturally occurring isolate, then the GoF experiment would not in general be under consideration.
Note the narrow scope of the question that such an experiment can answer. It cannot answer any of the following types of question:
1. Starting from a particular viral sequence, is a particular set of genetic changes necessary to create a phenotype of interest? This question is unanswerable in principle, because one can never make all possible genetic changes on a single starting genotype and rule out all alternative sets of changes [39].
2. Starting from a particular viral sequence, is a particular set of genetic changes the smallest set of changes on that genetic background sufficient to produce the phenotype? This question is unanswerable in practice for any set larger than about three mutations, because it is not possible to test phenotypically every one of the ~1012 combinations of three or more mutations, at least for whole-animal phenotypes like transmission that must be evaluated statistically in a number of animals for each genetic sequence. Thus even if each member of the set of changes is required along with the others to get the phenotype, there may be another set of (the same number of or fewer) changes that has not been tried in the experiment [39].
3. Is the set of mutations that produces the phenotype in the laboratory likely to happen in nature? This question can’t be answered by GoF experiments in the laboratory because viruses in nature are subject to many, possibly competing selective pressures and nonselective forces (e.g., population bottlenecks) that might produce a different trajectory of sequence changes from those that appear in the lab. Moreover, only a vanishingly small proportion of the viruses in nature have the genotype that forms the starting point of the experiment.
4. Is the set of mutations that produces the phenotype in the laboratory given a particular starting sequence likely to produce the phenotype when introduced into a viral strain with a different starting sequence? This would be predictable only if epistatic interactions were rare in influenza, such that the effect of a set of genetic changes on phenotype was relatively independent of genetic background. However, epistasis is pervasive in influenza [40, 41], including for phenotypes of interest such as and hemagglutinin receptor binding specificity [42, 43]. Thus, for example, the very mutations identified in GoF experiments on H5N1 isolates as conferring human receptor specificity on the viral hemagglutinin (HA) did not do so when introduced into a slightly different genetic background [43].
5. Does the set of mutations that confers ferret-to-ferret droplet transmissibility when introduced into a particular starting sequence also confer human-to-human transmissibility? Barring an unethical deliberate infection of humans with the virus resulting from GoF experiments in ferrets, and barring an accidental infection of humans, which would be potentially catastrophic, we will never know whether that particular virus’s transmission potential in ferrets translates to humans. Ferret droplet transmissibility is a very good, but not perfect predictor of human transmissibility for influenza [44]. At least one prominent influenza virologist has argued that the strains produced in GoF experiments in ferrets probably are not readily transmissible in humans [18], and others have said the same thing in public forums. If taken seriously, this uncertainty undermines the scientific and public health value of the experiments, because it undermines our ability to use the presence of GoF-identified mutations and phenotypes to gauge the level of risk presented by naturally occurring viruses [45].
4.1 What Scientific Questions Are of Greatest Importance for Pandemic Prevention and Response?
Each of the above classes of questions—those answerable by GoF experiments (in some cases by GoFRoC experiments) and those not answerable by such experiments—has legitimate scientific interest. But improving our ability to prevent and/or respond to novel influenza pandemics is the major public health goal that lends practical importance to advancing our scientific knowledge of the determinants of transmissibility in particular. Most importantly, effective targeting of efforts to prevent or mitigate pandemic threats partly depends on developing accurate predictive capabilities to estimate the magnitude of the threat posed by particular influenza A strains that we are observing in nonhuman reservoirs [40, 45]. Knowing that strains isolated from wild or domesticated avian or domesticated swine populations have genetic sequences or phenotypic traits (e.g., receptor binding specificity) predictive of the ability to transmit effectively in humans can motivate targeted countermeasures against such strains, such as culling of infected animals or strain-specific vaccine development. These two activities in particular depend on prioritizing which strains are most threatening, because it is impractical to cull all influenza-infected animals, and it is currently impractical to make vaccines except in a strainspecific fashion. Such strain-dependent prevention and mitigation activities are part of an overall pandemic preparedness portfolio including strengthening human respiratory disease surveillance and analytic capacity, stockpiling of non-strain-specific countermeasures such as antiviral drugs, personal protective equipment and ventilators, and a range of other public health measures whose usefulness is relatively independent of the strain of influenza that causes zoonotic cases or initiates a pandemic. But for animal culling and vaccine development (absent universal vaccines), prioritizing pandemic threats based on sequence and viral traits is important [40, 45, 46].
Many different types of scientific approaches can contribute to such prioritization efforts [7]. Comparison of naturally occurring influenza viruses, either by genetic sequence or by phenotypic traits or both, can help to categorize those sequences and traits that are characteristics of strains that are avian-adapted and poorly able to transmit in humans, compared to those that are well-adapted to human transmission. Such comparisons have been made for decades, and a number of traits and genetic sequence features (such as specific variants at specific amino acid sites in specific viral proteins) have been associated with human adaptation. Among the most important traits correlated with human adaptation are HA specificity for sialic acids found in the human upper airway (alpha-2,6 linked), a relatively low pH of HA activation, and human adaptation of the nucleoprotein-polymerase complex [40].
As the importance of these and other traits has become apparent from natural comparisons, the sequence variations correlated with these properties have been established through biochemical and other assays of single gene products or minigenomes (in the case of the nucleoprotein-polymerase complex) [47]. Such sequence changes can then be tested for their causal role in the individual traits, measurable in vitro, again without employing infectious virus [48–50]. Gain-of-function studies can be performed without employing strains with pandemic potential, for example, by taking a natural strain, introducing a change that abrogates one of the traits of interest, and then selecting revertants (so-called gain-loss-of-function) [51] or by doing gain-of-function experiments similar to GoFRoC but employing attenuated strains [52], strains of lower virulence, and strains for which there is greater population-wide immunity. All of these are alternatives to GoFRoC, and each is capable of informing public health efforts to prioritize pandemic threats from naturally occurring strains.
Importantly, such prioritization efforts are and will remain intrinsically imperfect. There have only been four influenza pandemics since the turn of the twentieth century, so our evolutionary models of what confers human-to-human transmissibility are constrained by being based on essentially four independent data points. The limitations of predicting pandemic potential were evident in 2009, when the strain that emerged and began spreading in humans arguably failed to meet the existing notions of human adaptation on all three key human-adaptive traits as understood at the time: early pandemic isolates’ HA receptor specificity was mixed human and avian; their HA pH of activation was outside the range of human-adapted viruses known before; and they lacked the sequence variant (lysine at PB2 amino acid 627) that had been thought necessary in the past for human adaptation. While subsequent evolution “corrected” the first two of these limitations on human-to-human transmission, the criteria had to be updated to accommodate the fact that what seemed necessary for pandemic potential in 2008 was violated by the early pandemic strain in 2009 [40]. While this limitation is surely a rationale for continuing to expand our scientific knowledge in this area, it more fundamentally points out that with only four modern pandemics to test our models of what is needed for a pandemic strain, no amount of further research will be able to make a highly reliable predictive model for pandemic potential—we simply lack the data (actual pandemics) to validate such models and therefore can never rely fully on these models, no matter how many more experimental inputs we add. This consideration heightens the importance of other approaches to pandemic preparedness, which are not dependent on correct predictions of the strain that will cause a pandemic, in combination with efforts to improve our predictive power. Table 1 lists a number of experimental and computational approaches to increasing our understanding of influenza pandemic risk determinants (top) and increasing our preparedness for pandemics more generally (bottom).
- admin
- Site Admin
- Posts: 36135
- Joined: Thu Aug 01, 2013 5:21 am
Re: U.S. government gave $3.7 million grant to Wuhan lab at
Part 2 of 2
5. What Do We Achieve, from a Public Health Perspective, with GoFRoC, that We Cannot Achieve with Safer Alternatives?
Given these considerations, no reasonable number of GoFRoC studies will give us enough data to be certain we know all the ways that influenza can achieve human-to-human transmission. Yet dramatic public health benefits have been claimed for GoFRoC experiments. Authors from the US CDC wrote in 2015 that prioritization of countermeasures against H7N9 strains in Cambodia that year had been possible uniquely thanks to the GoFRoC experiments that identified the importance of particular mutations in achieving human-to-human transmissibility [53]. These claims were exaggerated in the sense that every one of the mutations identified by those authors as generating concern about the Cambodian H7N9 strains, which had been identified in GoFRoC with H5N1, had been previously pinpointed in publications describing sequence analysis of existing strains and/or experimental approaches not constituting GoFRoC, such as binding studies of purified HA; the relevant experiments are listed in a table in Ref. [54]. While the GoFRoC studies in H5N1 added to the evidence base for the importance of these sites, there was nothing unique about the GoFRoC studies, and the same level of concern would have been appropriate purely on the basis of prior studies.
Similarly, participants in the recreation of the 1918 influenza A H1N1 strain asserted after the 2009 pandemic that this recreation had produced major public health benefits in the 2009 pandemic due to the understanding those experiments provided of H1N1 biology [5]. Yet the examples they cited, such as recognizing the likely protection enjoyed by elderly persons in 2009 who had been alive in 1918, in no way depended on the actual reconstruction of the 1918 virus. Sequence comparisons between the 2009 and 1918 viruses would have suggested the hypothesis, and serological assays of persons born before 1918 using the 2009 pandemic virus could—and did—test the hypothesis.
6. Conclusion
In summary, pandemic prediction, prevention, and mitigation are necessarily inexact sciences, where uncertainty remains mainly because we have so few pandemics to study. GoFRoC can add to the evidence base, but it cannot qualitatively change that evidence base. Empirically, the contribution of such studies to applied public health goals has been far more modest than claimed. Accounting for the unique risk posed by GoFRoC experiments as a factor in deciding whether to pursue them—as one should—shines a light on the narrowness of the scientific questions that they alone can answer. Vast advances in the most essential questions of influenza virology and in the public health goal of pandemic preparedness can be achieved without undertaking experiments that, if an accident occurs, could start a new pandemic. The consequences of such an accident should be enough to direct the attention of ambitious and public health-minded virologists toward these safe alternatives.
References
1. Duprex WP, Fouchier RA, Imperiale MJ, Lipsitch M, Relman DA (2015) Gain-of-function experiments: time for a real debate. Nat Rev Microbiol 13(1):58–64. https://doi.org/ 10.1038/nrmicro3405
2. Fouchier RA (2015) Studies on influenza virus transmission between ferrets: the public health risks revisited. MBio 6(1):e02560. https://doi. org/10.1128/mBio.02560-14
3. Butler D (2011) Fears grow over lab-bred flu. Nature 480(7378):421–422. https://doi.org/ 10.1038/480421a
4. Casadevall A, Howard D, Imperiale MJ (2014) An epistemological perspective on the value of gain-of-function experiments involving pathogens with pandemic potential. MBio 5(5): e01875. https://doi.org/10.1128/mBio. 01875-14
5. Taubenberger JK, Baltimore D, Doherty PC, Markel H, Morens DM, Webster RG, Wilson IA (2012) Reconstruction of the 1918 influenza virus: unexpected rewards from the past. MBio 3(5):e00201. https://doi.org/10. 1128/mBio.00201-12
6. Lipsitch M, Galvani A (2014) COMMENTARY: The case against ‘gain-of-function’ experiments: A reply to Fouchier & Kawaoka. CIDRAP June 19 http://www.cidrap.umn. edu/news-perspective/2014/06/commen tary-case-against-gain-function-experimentsreply- fouchier-kawaoka. Accessed 18 Nov 2014
7. Lipsitch M, Galvani AP (2014) Ethical alternatives to experiments with novel potential pandemic pathogens. PLoS Med 11(5):e1001646. https://doi.org/10.1371/journal.pmed. 1001646
8. Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, deWit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA (2012) Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336 (6088):1534–1541. https://doi.org/10. 1126/science.1213362
9. Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y (2012) Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486(7403):420–428. https://doi.org/10. 1038/nature10831
10. Sorrell EM, Wan H, Araya Y, Song H, Perez DR (2009) Minimal molecular constraints for respiratory droplet transmission of an avianhuman H9N2 influenza A virus. Proc Natl Acad Sci U S A 106(18):7565–7570. https:// doi.org/10.1073/pnas.0900877106
11. Kimble JB, Angel M, Wan H, Sutton TC, Finch C, Perez DR (2013) Alternative reassortment events leading to transmissible H9N1 influenza viruses in the ferret model. J Virol 88:66. https://doi.org/10.1128/JVI.02677- 13
12. Zhang Y, Zhang Q, Kong H, Jiang Y, Gao Y, Deng G, Shi J, Tian G, Liu L, Liu J, Guan Y, Bu Z, Chen H (2013) H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in Guinea pigs by respiratory droplet. Science 340(6139):1459–1463. https://doi.org/10. 1126/science.1229455
13. Sutton TC, Finch C, Shao H, Angel M, Chen H, Capua I, Cattoli G, Monne I, Perez DR (2014) Airborne transmission of highly pathogenic H7N1 influenza virus in ferrets. J Virol 88(12):6623–6635. https://doi.org/10. 1128/JVI.02765-13
14. Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A (2005) Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310(5745):77–80. https://doi.org/10. 1126/science.1119392
15. Enserink M (2004) Virology. tiptoeing around Pandora’s box. Science 305(5684):594–595. https://doi.org/10.1126/science.305.5684. 594
16. Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai le Q, Sedyaningsih ER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM (2006) Lack of transmission of H5N1 avianhuman reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A 103 (32):12121–12126. https://doi.org/10. 1073/pnas.0605134103
17. Maines TR, Chen LM, Belser JA, Van Hoeven N, Smith E, Donis RO, Tumpey TM, Katz JM (2011) Multiple genes contribute to the virulent phenotype observed in ferrets of an H5N1 influenza virus isolated from Thailand in 2004. Virology 413(2):226–230. https:// doi.org/10.1016/j.virol.2011.02.005
18. Palese P, Wang TT (2012) H5N1 influenza viruses: facts, not fear. Proc Natl Acad Sci U S A 109(7):2211–2213. https://doi.org/10. 1073/pnas.1121297109
19. Enserink M (2011) Infectious diseases. Controversial studies give a deadly flu virus wings. Science 334(6060):1192–1193. https://doi. org/10.1126/science.334.6060.1192
20. Fouchier RA, Garcia-Sastre A, Kawaoka Y, Barclay WS, Bouvier NM, Brown IH, Capua I, Chen H, Compans RW, Couch RB, Cox NJ, Doherty PC, Donis RO, Feldmann H, Guan Y, Katz J, Klenk HD, Kobinger G, Liu J, Liu X, Lowen A, Mettenleiter TC, Osterhaus AD, Palese P, Peiris JS, Perez DR, Richt JA, Schultz-Cherry S, Steel J, Subbarao K, Swayne DE, Takimoto T, Tashiro M, Taubenberger JK, Thomas PG, Tripp RA, Tumpey TM, Webby RJ, Webster RG (2012) Pause on avian flu transmission research. Science 335 (6067):400–401. https://doi.org/10.1126/ science.335.6067.400 10.1126/ science.1219412
21. Fouchier RA, Garcia-Sastre A, Kawaoka Y (2012) Pause on avian flu transmission studies. Nature 481(7382):443. https://doi.org/10. 1038/481443a
22. Fouchier RA, Garcia-Sastre A, Kawaoka Y (2013) H5N1 virus: transmission studies resume for avian flu. Nature 493(7434):609. https://doi.org/10.1038/nature11858
23. Fouchier RA, Garcia-Sastre A, Kawaoka Y, Barclay WS, Bouvier NM, Brown IH, Capua I, Chen H, Compans RW, Couch RB, Cox NJ, Doherty PC, Donis RO, Feldmann H, Guan Y, Katz JM, Kiselev OI, Klenk HD, Kobinger G, Liu J, Liu X, Lowen A, Mettenleiter TC, Osterhaus AD, Palese P, Peiris JS, Perez DR, Richt JA, Schultz-Cherry S, Steel J, Subbarao K, Swayne DE, Takimoto T, Tashiro M, Taubenberger JK, Thomas PG, Tripp RA, Tumpey TM, Webby RJ, Webster RG (2013) Transmission studies resume for avian flu. Science 339 (6119):520–521. https://doi.org/10.1126/ science.1235140
24. Fouchier R, Osterhaus AB, Steinbruner J, Yuen KY, Henderson DA, Klotz L, Sylvester E, Taubenberger JK, Ebright RH, Heymann DL (2012) Preventing pandemics: the fight over flu. Nature 481(7381):257–259. https://doi. org/10.1038/481257a
25. Klotz LC, Sylvester EJ (2012) The unacceptable risks of a man-made pandemic. Bulletin of the Atomic Scientists web edition: http://the bulletin.org/unacceptable-risks-man-madepandemic
26. Lipsitch M, Plotkin JB, Simonsen L, Bloom B (2012) Evolution, safety, and highly pathogenic influenza viruses. Science 336 (6088):1529–1531. https://doi.org/10. 1126/science.1223204
27. Kaiser J (2014) U.S. halts two dozen risky virus studies. Science 346(6208):404. https://doi. org/10.1126/science.346.6208.404
28. Council IoMaNR (2015) Potential risks and benefits of gain-of-function research: summary of a workshop. National Academies Press. http://dels.nas.edu/Workshop-Summary/ Potential-Risks-Benefits-Gain/21666, Washington, DC. https://doi.org/10.17226/ 21666
29. National Academies of Sciences E, and Medicine (2016) Gain-of-function research: summary of the second symposium, March 10–11, 2016. National Academies Press. https://www.nap.edu/catalog/23484/gainof- function-research-summary-of-the-secondsymposium- march, Washington, DC. https:// doi.org/10.17226/23484
30. Lipsitch M (2014) Can limited scientific value of potential pandemic pathogen experiments justify the risks? MBio 5(5):e02008. https:// doi.org/10.1128/mBio.02008-14
31. Evans NG, Lipsitch M, Levinson M (2015) The ethics of biosafety considerations in gainof- function research resulting in the creation of potential pandemic pathogens. J Med Ethics 41(11):901–908. https://doi.org/10.1136/ medethics-2014-102619
32. Kimman TG, Smit E, Klein MR (2008) Evidence-based biosafety: a review of the principles and effectiveness of microbiological containment measures. Clin Microbiol Rev 21 (3):403–425. https://doi.org/10.1128/ CMR.00014-08
33. Harding A, Byers K (2006) Epidemiology of laboratory-associated infections. In: Fleming D, Hunt D (eds) Biological safety. ASM Press, Washington, DC, pp 53–77. https://doi.org/10.1128/9781555815899. ch4
34. Lipsitch M, Inglesby TV (2014) Moratorium on research intended to create novel potential pandemic pathogens. MBio 5(6):e02366. https://doi.org/10.1128/mBio.02366-14
35. Centers for Disease Control and Prevention (2014) Report on the potential exposure to anthrax. http://www.cdc.gov/about/pdf/ lab-safety/Final_Anthrax_Report.pdf
36. Grady D, McNeil DG (2014) Ebola sample is mishandled at C.D.C. Lab in latest error. New York Times, p A1. http://www.nytimes. com/2014/12/25/health/cdc-ebola-errorin- lab-may-have-exposed-technician-to-virus. html
37. CDC (2014) Report on the inadvertent crosscontamination and shipment of a laboratory specimen with influenza virus H5N1. http:// http://www.cdc.gov/about/pdf/lab-safety/investi gationcdch5n1contaminationeventaugust 15pdf
38. Lipsitch M, Inglesby TV (2015) Reply to “studies on influenza virus transmission between ferrets: the public health risks revisited”. MBio 6(1):e00041. https://doi.org/10. 1128/mBio.00041-15
39. Lipsitch M (2014) Lack of statistical support for claim of “minimal set of substitutions” and limitations of the Ferret Model for human transmissibility [online comment on linster et al. identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus]. Cell 127:329–339 http://www.cell.com/cell/com ments/S0092-8674(14)00281-5
40. Lipsitch M, Barclay W, Raman R, Russell CJ, Belser JA, Cobey S, Kasson PM, Lloyd-Smith JO, Maurer-Stroh S, Riley S, Beauchemin CA, Bedford T, Friedrich TC, Handel A, Herfst S, Murcia PR, Roche B, Wilke CO, Russell CA (2016) Viral factors in influenza pandemic risk assessment. eLife 5:e18491. https://doi.org/ 10.7554/eLife.18491
41. Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB (2011) Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS Genet 7(2):e1001301. https://doi. org/10.1371/journal.pgen.1001301
42. Tharakaraman K, Jayaraman A, Raman R, Viswanathan K, Stebbins NW, Johnson D, Shriver Z, Sasisekharan V, Sasisekharan R (2013) Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell 153 (7):1486–1493. https://doi.org/10.1016/j. cell.2013.05.034
43. Tharakaraman K, Raman R, Viswanathan K, Stebbins NW, Jayaraman A, Krishnan A, Sasisekharan V, Sasisekharan R (2013) Structural determinants for naturally evolving H5N1 hemagglutinin to switch its receptor specificity. Cell 153(7):1475–1485. https:// doi.org/10.1016/j.cell.2013.05.035
44. Buhnerkempe MG, Gostic K, Park M, Ahsan P, Belser JA, Lloyd-Smith JO (2015) Mapping influenza transmission in the ferret model to transmission in humans. eLife 4:e07969. https://doi.org/10.7554/eLife.07969
45. Russell CA, Kasson PM, Donis RO, Riley S, Dunbar J, Rambaut A, Asher J, Burke S, Davis CT, Garten RJ, Gnanakaran S, Hay SI, Herfst S, Lewis NS, Lloyd-Smith JO, Macken CA, Maurer-Stroh S, Neuhaus E, Parrish CR, Pepin KM, Shepard SS, Smith DL, Suarez DL, Trock SC, Widdowson MA, George DB, Lipsitch M, Bloom JD (2014) Improving pandemic influenza risk assessment. eLife 3: e03883. https://doi.org/10.7554/eLife. 03883
46. Trock SC, Burke SA, Cox NJ (2012) Development of an influenza virologic risk assessment tool. Avian Dis 56(4 Suppl):1058–1061
47. Moncorge O, Mura M, Barclay WS (2010) Evidence for avian and human host cell factors that affect the activity of influenza virus polymerase. J Virol 84(19):9978–9986. https:// doi.org/10.1128/JVI.01134-10
48. Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, McCauley JW, Locher K, Walker PA, Collins PJ, Kawaoka Y, Skehel JJ, Gamblin SJ (2013) Receptor binding by a ferrettransmissible H5 avian influenza virus. Nature 497(7449):392–396. https://doi.org/10. 1038/nature12144
49. Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA (2006) Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol 355 (5):1143–1155. https://doi.org/10.1016/j. jmb.2005.11.002
50. Russier M, Yang G, Marinova-Petkova A, Vogel P, Kaplan BS, Webby RJ, Russell CJ (2017) H1N1 influenza viruses varying widely in hemagglutinin stability transmit efficiently from swine to swine and to ferrets. PLoS Pathog 13(3):e1006276. https://doi.org/10. 1371/journal.ppat.1006276
51. Lakdawala SS, Jayaraman A, Halpin RA, Lamirande EW, Shih AR, Stockwell TB, Lin X, Simenauer A, Hanson CT, Vogel L, Paskel M, Minai M, Moore I, Orandle M, Das SR, Wentworth DE, Sasisekharan R, Subbarao K (2015) The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 526:122. https://doi.org/10.1038/ nature15379
52. Krenn BM, Egorov A, Romanovskaya- Romanko E, Wolschek M, Nakowitsch S, Ruthsatz T, Kiefmann B, Morokutti A, Humer J, Geiler J, Cinatl J, Michaelis M, Wressnigg N, Sturlan S, Ferko B, Batishchev OV, Indenbom AV, Zhu R, Kastner M, Hinterdorfer P, Kiselev O, Muster T, Romanova J (2011) Single HA2 mutation increases the infectivity and immunogenicity of a live attenuated H5N1 intranasal influenza vaccine candidate lacking NS1. PLoS One 6(4): e18577. https://doi.org/10.1371/journal. pone.0018577
53. Davis CT, Chen LM, Pappas C, Stevens J, Tumpey TM, Gubareva LV, Katz JM, Villanueva JM, Donis RO, Cox NJ (2014) Use of highly pathogenic avian influenza A(H5N1) gain-of-function studies for molecular-based surveillance and pandemic preparedness. MBio 5(6):e02431. https://doi.org/10. 1128/mBio.02431-14
54. Lipsitch M (2016) Comment on “gain-offunction research and the relevance to clinical practice”. J Infect Dis 214(8):1284–1285. https://doi.org/10.1093/infdis/jiw348
55. Jimenez-Alberto A, Alvarado-Facundo E, Ribas-Aparicio RM, Castelan-Vega JA (2013) Analysis of adaptation mutants in the hemagglutinin of the influenza A(H1N1)pdm09 virus. PLoS One 8(7):e70005. https://doi. org/10.1371/journal.pone.0070005
56. Larsson P, Kasson PM (2013) Lipid tail protrusion in simulations predicts fusogenic activity of influenza fusion peptide mutants and conformational models. PLoS Comput Biol 9(3): e1002950. https://doi.org/10.1371/journal. pcbi.1002950
57. Ripoll DR, Khavrutskii IV, Chaudhury S, Liu J, Kuschner RA, Wallqvist A, Reifman J (2012) Quantitative predictions of binding free energy changes in drug-resistant influenza neuraminidase. PLoS Comput Biol 8(8):e1002665. https://doi.org/10.1371/journal.pcbi. 1002665
58. Shelton H, Roberts KL, Molesti E, Temperton N, Barclay WS (2013) Mutations in haemagglutinin that affect receptor binding and pH stability increase replication of a PR8 influenza virus with H5 HA in the upper respiratory tract of ferrets and may contribute to transmissibility. J Gen Virol 94 (Pt 6):1220–1229. https://doi.org/10. 1099/vir.0.050526-0
59. Shelton H, Ayora-Talavera G, Ren J, Loureiro S, Pickles RJ, Barclay WS, Jones IM (2011) Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J Virol 85(4):1875–1880. https://doi.org/10. 1128/JVI.01822-10
60. Gong LI, Suchard MA, Bloom JD (2013) Stability-mediated epistasis constrains the evolution of an influenza protein. eLife 2:e00631. https://doi.org/10.7554/eLife.00631
61. Cauldwell AV, Moncorge O, Barclay WS (2013) Unstable polymerase-nucleoprotein interaction is not responsible for avian influenza virus polymerase restriction in human cells. J Virol 87(2):1278–1284. https://doi. org/10.1128/JVI.02597-12
62. Tamuri AU, Dos Reis M, Hay AJ, Goldstein RA (2009) Identifying changes in selective constraints: host shifts in influenza. PLoS Comput Biol 5(11):e1000564. https://doi. org/10.1371/journal.pcbi.1000564
63. dos Reis M, Tamuri AU, Hay AJ, Goldstein RA (2011) Charting the host adaptation of influenza viruses. Mol Biol Evol 28(6):1755–1767. https://doi.org/10.1093/molbev/msq317
64. Jonges M, Meijer A, Fouchier RA, Koch G, Li J, Pan JC, Chen H, Shu YL, Koopmans MP (2013) Guiding outbreak management by the use of influenza A(H7Nx) virus sequence analysis. Euro Surveill 18(16):20460
65. Wong EH, Smith DK, Rabadan R, Peiris M, Poon LL (2010) Codon usage bias and the evolution of influenza A viruses. Codon usage biases of influenza virus. BMC Evol Biol 10:253. https://doi.org/10.1186/1471- 2148-10-253
66. Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H (2013) H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341 (6144):410–414. https://doi.org/10.1126/ science.1240532
67. Gustin KM, Katz JM, Tumpey TM, Maines TR (2013) Comparison of the levels of infectious virus in respirable aerosols exhaled by ferrets infected with influenza viruses exhibiting diverse transmissibility phenotypes. J Virol 87 (14):7864–7873. https://doi.org/10.1128/ JVI.00719-13
68. Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, Garcia-Sastre A, Palese P, Hai R (2011) The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the Guinea pig model. J Virol 85(21):11235–11241. https:// doi.org/10.1128/JVI.05794-11
69. Yen HL, Liang CH, Wu CY, Forrest HL, Ferguson A, Choy KT, Jones J, Wong DD, Cheung PP, Hsu CH, Li OT, Yuen KM, Chan RW, Poon LL, Chan MC, Nicholls JM, Krauss S, Wong CH, Guan Y, Webster RG, Webby RJ, Peiris M (2011) Hemagglutininneuraminidase balance confers respiratorydroplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A 108(34):14264–14269. https://doi.org/10.1073/pnas.1111000108
70. Krammer F, Palese P (2013) Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3(5):521–530. https:// doi.org/10.1016/j.coviro.2013.07.007
71. Wei CJ, Yassine HM, McTamney PM, Gall JG, Whittle JR, Boyington JC, Nabel GJ (2012) Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Sci Transl Med 4(147):147ra114. https://doi.org/10.1126/scitranslmed. 3004273
72. Schmitz N, Beerli RR, Bauer M, Jegerlehner A, Dietmeier K, Maudrich M, Pumpens P, Saudan P, Bachmann MF (2012) Universal vaccine against influenza virus: linking TLR signaling to anti-viral protection. Eur J Immunol 42(4):863–869. https://doi.org/10. 1002/eji.201041225
73. Hughes B, Hayden F, Perikov Y, Hombach J, Tam JS (2012) Report of the 5th meeting on influenza vaccines that induce broad spectrum and long-lasting immune responses, World Health Organization, Geneva, 16–- 17 November 2011. Vaccine 30 (47):6612–6622. https://doi.org/10.1016/j. vaccine.2012.08.073
74. Doyle TM, Jaentschke B, Van Domselaar G, Hashem AM, Farnsworth A, Forbes NE, Li C, Wang J, He R, Brown EG, Li X (2013) The universal epitope of influenza A viral neuraminidase fundamentally contributes to enzyme activity and viral replication. J Biol Chem 288(25):18283–18289. https://doi. org/10.1074/jbc.M113.468884
75. Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, Digard P, Anttila V, Baillie JK, Walsh TS, Hume DA, Palotie A, Xue Y, Colonna V, Tyler-Smith C, Dunning J, Gordon SB, Smyth RL, Openshaw PJ, Dougan G, Brass AL, Kellam P (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484(7395):519–523. https:// doi.org/10.1038/nature10921
76. Dormitzer PR, Suphaphiphat P, Gibson DG, Wentworth DE, Stockwell TB, Algire MA, Alperovich N, Barro M, Brown DM, Craig S, Dattilo BM, Denisova EA, De Souza I, Eickmann M, Dugan VG, Ferrari A, Gomila RC, Han L, Judge C, Mane S, Matrosovich M, Merryman C, Palladino G, Palmer GA, Spencer T, Strecker T, Trusheim H, Uhlendorff J, Wen Y, Yee AC, Zaveri J, Zhou B, Becker S, Donabedian A, Mason PW, Glass JI, Rappuoli R, Venter JC (2013) Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci Transl Med 5(185):185ra168. https://doi. org/10.1126/scitranslmed.3006368
______________
Notes:
Yohei Yamauchi (ed.), Influenza Virus: Methods and Protocols, Methods in Molecular Biology, vol. 1836, https://doi.org/10.1007/978-1-4939-8678-1_29, © Springer Science+Business Media, LLC, part of Springer Nature 2018
5. What Do We Achieve, from a Public Health Perspective, with GoFRoC, that We Cannot Achieve with Safer Alternatives?
Given these considerations, no reasonable number of GoFRoC studies will give us enough data to be certain we know all the ways that influenza can achieve human-to-human transmission. Yet dramatic public health benefits have been claimed for GoFRoC experiments. Authors from the US CDC wrote in 2015 that prioritization of countermeasures against H7N9 strains in Cambodia that year had been possible uniquely thanks to the GoFRoC experiments that identified the importance of particular mutations in achieving human-to-human transmissibility [53]. These claims were exaggerated in the sense that every one of the mutations identified by those authors as generating concern about the Cambodian H7N9 strains, which had been identified in GoFRoC with H5N1, had been previously pinpointed in publications describing sequence analysis of existing strains and/or experimental approaches not constituting GoFRoC, such as binding studies of purified HA; the relevant experiments are listed in a table in Ref. [54]. While the GoFRoC studies in H5N1 added to the evidence base for the importance of these sites, there was nothing unique about the GoFRoC studies, and the same level of concern would have been appropriate purely on the basis of prior studies.
Similarly, participants in the recreation of the 1918 influenza A H1N1 strain asserted after the 2009 pandemic that this recreation had produced major public health benefits in the 2009 pandemic due to the understanding those experiments provided of H1N1 biology [5]. Yet the examples they cited, such as recognizing the likely protection enjoyed by elderly persons in 2009 who had been alive in 1918, in no way depended on the actual reconstruction of the 1918 virus. Sequence comparisons between the 2009 and 1918 viruses would have suggested the hypothesis, and serological assays of persons born before 1918 using the 2009 pandemic virus could—and did—test the hypothesis.
Table 1: Scientific approaches that are scientifically valuable for understanding influenza virus determinants of pandemic potential and more generally for improving pandemic preparedness, but do not risk the creation of potential pandemic pathogens (PPPs)
Approach / Examples / Scientific benefits
Molecular dynamic modeling of influenza proteins and interactions with inhibitors and receptors / Analysis of adaptive changes in hemagglutinin (HA) of H1N1pdm [55], lipid tail protrusion as a determinant of HA membrane fusion [56], and identifying determinants of inhibitor-resistant neuraminidase [57] / Biophysical basis for complex phenotypes
In vitro studies of specific properties required for human adaptation, using single proteins / Studies of H5 or H7 receptor binding to mammalian vs. human sialic acids [42, 43]; Studies of genetic determinants of optimal pH of fusion by comparing properties of natural isolates [58] / Higher throughput than in vivo studies; can study more sequences and define motifs required for binding, beyond individual mutations; ability to test generality of hypothesized determinants [59]
In vitro studies of genetic interactions between loci in one or several viral proteins using replication-incompetent viruses / Studies of epistatic interactions in nucleoprotein [60] or between nucleoprotein and polymerase [61] using in vitro expression of markers and stability measurements of proteins / Higher throughput; ability to link structure to function; ability to test combinations of mutations
Sequence database comparisons of genetic properties of human- and avian-adapted viruses / Identify amino acid markers of host adaptation, and quantify the extent of adaptation to a particular host [62, 63]; Search for markers of human adaptation (established in earlier studies without PPP production) in H7N9 viruses [64] / Very high throughput; future studies could use novel analytic methods [65] to systematically identify new markers associated with human adaptation, which could then be tested experimentally; focus on naturally viable mutations
Sequence and in vitro phenotypic comparisons of human seasonal influenza isolates, zoonotic isolates from infected humans, and avian isolates / Comparison of human and avian isolates of H7N9 [66] Comparison of viral shedding in ferrets of human seasonal and pandemic vs. avian H5N1 viruses [67] / Focus on naturally viable variants; higher throughput; ability to test a wide range of phenotypes
Experimental production and testing in animal transmission models of reassortants or mutants of seasonal influenza to identify genetic components required for transmissibility, maintaining surface proteins to which human immunity exists / Replacing M segment of H3N2 and H1N1 strains with one from H1N1pdm to assess effect on guinea pig transmission [68]; Ferret transmission assays of recombinant H1N2swine x H1N1pdm viruses to assess role of HA-NA balance [69] / Human transmissibility of parent viruses provides “natural” validation of animal model
Loss+gain-of-function in existing viruses to which humans already have significant immunity / Showing that the soft palate is an important site of HA adaptation to airborne transmission in ferrets, using a loss-of-function engineered mutant in H1N1pdm [51] / Gain-of-function studied without creating a PPP
Universal or broadly neutralizing influenza vaccine research / Hemagglutinin stalk vaccines [70, 71]; enhancing responses to conserved proteins [72]; T cell vaccination and improved adjuvants [73]; targeting universal neuraminidase epitopes [74] / Successful vaccine could eliminate need for rapid production of pandemicspecific vaccine and seasonal revaccination; complementary technology to other approaches
Studies of host factors using naturally occurring viruses / Identification of host factors restricting pathogenicity in animal models, in vitro and via human genetics [75] / Potential therapeutic targets identified
Accelerating vaccine production / Sequence-based design and cell culture manufacture of influenza vaccine [76] / More rapid manufacture
Modified from [7]
6. Conclusion
In summary, pandemic prediction, prevention, and mitigation are necessarily inexact sciences, where uncertainty remains mainly because we have so few pandemics to study. GoFRoC can add to the evidence base, but it cannot qualitatively change that evidence base. Empirically, the contribution of such studies to applied public health goals has been far more modest than claimed. Accounting for the unique risk posed by GoFRoC experiments as a factor in deciding whether to pursue them—as one should—shines a light on the narrowness of the scientific questions that they alone can answer. Vast advances in the most essential questions of influenza virology and in the public health goal of pandemic preparedness can be achieved without undertaking experiments that, if an accident occurs, could start a new pandemic. The consequences of such an accident should be enough to direct the attention of ambitious and public health-minded virologists toward these safe alternatives.
References
1. Duprex WP, Fouchier RA, Imperiale MJ, Lipsitch M, Relman DA (2015) Gain-of-function experiments: time for a real debate. Nat Rev Microbiol 13(1):58–64. https://doi.org/ 10.1038/nrmicro3405
2. Fouchier RA (2015) Studies on influenza virus transmission between ferrets: the public health risks revisited. MBio 6(1):e02560. https://doi. org/10.1128/mBio.02560-14
3. Butler D (2011) Fears grow over lab-bred flu. Nature 480(7378):421–422. https://doi.org/ 10.1038/480421a
4. Casadevall A, Howard D, Imperiale MJ (2014) An epistemological perspective on the value of gain-of-function experiments involving pathogens with pandemic potential. MBio 5(5): e01875. https://doi.org/10.1128/mBio. 01875-14
5. Taubenberger JK, Baltimore D, Doherty PC, Markel H, Morens DM, Webster RG, Wilson IA (2012) Reconstruction of the 1918 influenza virus: unexpected rewards from the past. MBio 3(5):e00201. https://doi.org/10. 1128/mBio.00201-12
6. Lipsitch M, Galvani A (2014) COMMENTARY: The case against ‘gain-of-function’ experiments: A reply to Fouchier & Kawaoka. CIDRAP June 19 http://www.cidrap.umn. edu/news-perspective/2014/06/commen tary-case-against-gain-function-experimentsreply- fouchier-kawaoka. Accessed 18 Nov 2014
7. Lipsitch M, Galvani AP (2014) Ethical alternatives to experiments with novel potential pandemic pathogens. PLoS Med 11(5):e1001646. https://doi.org/10.1371/journal.pmed. 1001646
8. Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, deWit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA (2012) Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336 (6088):1534–1541. https://doi.org/10. 1126/science.1213362
9. Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y (2012) Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486(7403):420–428. https://doi.org/10. 1038/nature10831
10. Sorrell EM, Wan H, Araya Y, Song H, Perez DR (2009) Minimal molecular constraints for respiratory droplet transmission of an avianhuman H9N2 influenza A virus. Proc Natl Acad Sci U S A 106(18):7565–7570. https:// doi.org/10.1073/pnas.0900877106
11. Kimble JB, Angel M, Wan H, Sutton TC, Finch C, Perez DR (2013) Alternative reassortment events leading to transmissible H9N1 influenza viruses in the ferret model. J Virol 88:66. https://doi.org/10.1128/JVI.02677- 13
12. Zhang Y, Zhang Q, Kong H, Jiang Y, Gao Y, Deng G, Shi J, Tian G, Liu L, Liu J, Guan Y, Bu Z, Chen H (2013) H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in Guinea pigs by respiratory droplet. Science 340(6139):1459–1463. https://doi.org/10. 1126/science.1229455
13. Sutton TC, Finch C, Shao H, Angel M, Chen H, Capua I, Cattoli G, Monne I, Perez DR (2014) Airborne transmission of highly pathogenic H7N1 influenza virus in ferrets. J Virol 88(12):6623–6635. https://doi.org/10. 1128/JVI.02765-13
14. Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A (2005) Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310(5745):77–80. https://doi.org/10. 1126/science.1119392
15. Enserink M (2004) Virology. tiptoeing around Pandora’s box. Science 305(5684):594–595. https://doi.org/10.1126/science.305.5684. 594
16. Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai le Q, Sedyaningsih ER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM (2006) Lack of transmission of H5N1 avianhuman reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A 103 (32):12121–12126. https://doi.org/10. 1073/pnas.0605134103
17. Maines TR, Chen LM, Belser JA, Van Hoeven N, Smith E, Donis RO, Tumpey TM, Katz JM (2011) Multiple genes contribute to the virulent phenotype observed in ferrets of an H5N1 influenza virus isolated from Thailand in 2004. Virology 413(2):226–230. https:// doi.org/10.1016/j.virol.2011.02.005
18. Palese P, Wang TT (2012) H5N1 influenza viruses: facts, not fear. Proc Natl Acad Sci U S A 109(7):2211–2213. https://doi.org/10. 1073/pnas.1121297109
19. Enserink M (2011) Infectious diseases. Controversial studies give a deadly flu virus wings. Science 334(6060):1192–1193. https://doi. org/10.1126/science.334.6060.1192
20. Fouchier RA, Garcia-Sastre A, Kawaoka Y, Barclay WS, Bouvier NM, Brown IH, Capua I, Chen H, Compans RW, Couch RB, Cox NJ, Doherty PC, Donis RO, Feldmann H, Guan Y, Katz J, Klenk HD, Kobinger G, Liu J, Liu X, Lowen A, Mettenleiter TC, Osterhaus AD, Palese P, Peiris JS, Perez DR, Richt JA, Schultz-Cherry S, Steel J, Subbarao K, Swayne DE, Takimoto T, Tashiro M, Taubenberger JK, Thomas PG, Tripp RA, Tumpey TM, Webby RJ, Webster RG (2012) Pause on avian flu transmission research. Science 335 (6067):400–401. https://doi.org/10.1126/ science.335.6067.400 10.1126/ science.1219412
21. Fouchier RA, Garcia-Sastre A, Kawaoka Y (2012) Pause on avian flu transmission studies. Nature 481(7382):443. https://doi.org/10. 1038/481443a
22. Fouchier RA, Garcia-Sastre A, Kawaoka Y (2013) H5N1 virus: transmission studies resume for avian flu. Nature 493(7434):609. https://doi.org/10.1038/nature11858
23. Fouchier RA, Garcia-Sastre A, Kawaoka Y, Barclay WS, Bouvier NM, Brown IH, Capua I, Chen H, Compans RW, Couch RB, Cox NJ, Doherty PC, Donis RO, Feldmann H, Guan Y, Katz JM, Kiselev OI, Klenk HD, Kobinger G, Liu J, Liu X, Lowen A, Mettenleiter TC, Osterhaus AD, Palese P, Peiris JS, Perez DR, Richt JA, Schultz-Cherry S, Steel J, Subbarao K, Swayne DE, Takimoto T, Tashiro M, Taubenberger JK, Thomas PG, Tripp RA, Tumpey TM, Webby RJ, Webster RG (2013) Transmission studies resume for avian flu. Science 339 (6119):520–521. https://doi.org/10.1126/ science.1235140
24. Fouchier R, Osterhaus AB, Steinbruner J, Yuen KY, Henderson DA, Klotz L, Sylvester E, Taubenberger JK, Ebright RH, Heymann DL (2012) Preventing pandemics: the fight over flu. Nature 481(7381):257–259. https://doi. org/10.1038/481257a
25. Klotz LC, Sylvester EJ (2012) The unacceptable risks of a man-made pandemic. Bulletin of the Atomic Scientists web edition: http://the bulletin.org/unacceptable-risks-man-madepandemic
26. Lipsitch M, Plotkin JB, Simonsen L, Bloom B (2012) Evolution, safety, and highly pathogenic influenza viruses. Science 336 (6088):1529–1531. https://doi.org/10. 1126/science.1223204
27. Kaiser J (2014) U.S. halts two dozen risky virus studies. Science 346(6208):404. https://doi. org/10.1126/science.346.6208.404
28. Council IoMaNR (2015) Potential risks and benefits of gain-of-function research: summary of a workshop. National Academies Press. http://dels.nas.edu/Workshop-Summary/ Potential-Risks-Benefits-Gain/21666, Washington, DC. https://doi.org/10.17226/ 21666
29. National Academies of Sciences E, and Medicine (2016) Gain-of-function research: summary of the second symposium, March 10–11, 2016. National Academies Press. https://www.nap.edu/catalog/23484/gainof- function-research-summary-of-the-secondsymposium- march, Washington, DC. https:// doi.org/10.17226/23484
30. Lipsitch M (2014) Can limited scientific value of potential pandemic pathogen experiments justify the risks? MBio 5(5):e02008. https:// doi.org/10.1128/mBio.02008-14
31. Evans NG, Lipsitch M, Levinson M (2015) The ethics of biosafety considerations in gainof- function research resulting in the creation of potential pandemic pathogens. J Med Ethics 41(11):901–908. https://doi.org/10.1136/ medethics-2014-102619
32. Kimman TG, Smit E, Klein MR (2008) Evidence-based biosafety: a review of the principles and effectiveness of microbiological containment measures. Clin Microbiol Rev 21 (3):403–425. https://doi.org/10.1128/ CMR.00014-08
33. Harding A, Byers K (2006) Epidemiology of laboratory-associated infections. In: Fleming D, Hunt D (eds) Biological safety. ASM Press, Washington, DC, pp 53–77. https://doi.org/10.1128/9781555815899. ch4
34. Lipsitch M, Inglesby TV (2014) Moratorium on research intended to create novel potential pandemic pathogens. MBio 5(6):e02366. https://doi.org/10.1128/mBio.02366-14
35. Centers for Disease Control and Prevention (2014) Report on the potential exposure to anthrax. http://www.cdc.gov/about/pdf/ lab-safety/Final_Anthrax_Report.pdf
36. Grady D, McNeil DG (2014) Ebola sample is mishandled at C.D.C. Lab in latest error. New York Times, p A1. http://www.nytimes. com/2014/12/25/health/cdc-ebola-errorin- lab-may-have-exposed-technician-to-virus. html
37. CDC (2014) Report on the inadvertent crosscontamination and shipment of a laboratory specimen with influenza virus H5N1. http:// http://www.cdc.gov/about/pdf/lab-safety/investi gationcdch5n1contaminationeventaugust 15pdf
38. Lipsitch M, Inglesby TV (2015) Reply to “studies on influenza virus transmission between ferrets: the public health risks revisited”. MBio 6(1):e00041. https://doi.org/10. 1128/mBio.00041-15
39. Lipsitch M (2014) Lack of statistical support for claim of “minimal set of substitutions” and limitations of the Ferret Model for human transmissibility [online comment on linster et al. identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus]. Cell 127:329–339 http://www.cell.com/cell/com ments/S0092-8674(14)00281-5
40. Lipsitch M, Barclay W, Raman R, Russell CJ, Belser JA, Cobey S, Kasson PM, Lloyd-Smith JO, Maurer-Stroh S, Riley S, Beauchemin CA, Bedford T, Friedrich TC, Handel A, Herfst S, Murcia PR, Roche B, Wilke CO, Russell CA (2016) Viral factors in influenza pandemic risk assessment. eLife 5:e18491. https://doi.org/ 10.7554/eLife.18491
41. Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB (2011) Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS Genet 7(2):e1001301. https://doi. org/10.1371/journal.pgen.1001301
42. Tharakaraman K, Jayaraman A, Raman R, Viswanathan K, Stebbins NW, Johnson D, Shriver Z, Sasisekharan V, Sasisekharan R (2013) Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell 153 (7):1486–1493. https://doi.org/10.1016/j. cell.2013.05.034
43. Tharakaraman K, Raman R, Viswanathan K, Stebbins NW, Jayaraman A, Krishnan A, Sasisekharan V, Sasisekharan R (2013) Structural determinants for naturally evolving H5N1 hemagglutinin to switch its receptor specificity. Cell 153(7):1475–1485. https:// doi.org/10.1016/j.cell.2013.05.035
44. Buhnerkempe MG, Gostic K, Park M, Ahsan P, Belser JA, Lloyd-Smith JO (2015) Mapping influenza transmission in the ferret model to transmission in humans. eLife 4:e07969. https://doi.org/10.7554/eLife.07969
45. Russell CA, Kasson PM, Donis RO, Riley S, Dunbar J, Rambaut A, Asher J, Burke S, Davis CT, Garten RJ, Gnanakaran S, Hay SI, Herfst S, Lewis NS, Lloyd-Smith JO, Macken CA, Maurer-Stroh S, Neuhaus E, Parrish CR, Pepin KM, Shepard SS, Smith DL, Suarez DL, Trock SC, Widdowson MA, George DB, Lipsitch M, Bloom JD (2014) Improving pandemic influenza risk assessment. eLife 3: e03883. https://doi.org/10.7554/eLife. 03883
46. Trock SC, Burke SA, Cox NJ (2012) Development of an influenza virologic risk assessment tool. Avian Dis 56(4 Suppl):1058–1061
47. Moncorge O, Mura M, Barclay WS (2010) Evidence for avian and human host cell factors that affect the activity of influenza virus polymerase. J Virol 84(19):9978–9986. https:// doi.org/10.1128/JVI.01134-10
48. Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, McCauley JW, Locher K, Walker PA, Collins PJ, Kawaoka Y, Skehel JJ, Gamblin SJ (2013) Receptor binding by a ferrettransmissible H5 avian influenza virus. Nature 497(7449):392–396. https://doi.org/10. 1038/nature12144
49. Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA (2006) Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol 355 (5):1143–1155. https://doi.org/10.1016/j. jmb.2005.11.002
50. Russier M, Yang G, Marinova-Petkova A, Vogel P, Kaplan BS, Webby RJ, Russell CJ (2017) H1N1 influenza viruses varying widely in hemagglutinin stability transmit efficiently from swine to swine and to ferrets. PLoS Pathog 13(3):e1006276. https://doi.org/10. 1371/journal.ppat.1006276
51. Lakdawala SS, Jayaraman A, Halpin RA, Lamirande EW, Shih AR, Stockwell TB, Lin X, Simenauer A, Hanson CT, Vogel L, Paskel M, Minai M, Moore I, Orandle M, Das SR, Wentworth DE, Sasisekharan R, Subbarao K (2015) The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 526:122. https://doi.org/10.1038/ nature15379
52. Krenn BM, Egorov A, Romanovskaya- Romanko E, Wolschek M, Nakowitsch S, Ruthsatz T, Kiefmann B, Morokutti A, Humer J, Geiler J, Cinatl J, Michaelis M, Wressnigg N, Sturlan S, Ferko B, Batishchev OV, Indenbom AV, Zhu R, Kastner M, Hinterdorfer P, Kiselev O, Muster T, Romanova J (2011) Single HA2 mutation increases the infectivity and immunogenicity of a live attenuated H5N1 intranasal influenza vaccine candidate lacking NS1. PLoS One 6(4): e18577. https://doi.org/10.1371/journal. pone.0018577
53. Davis CT, Chen LM, Pappas C, Stevens J, Tumpey TM, Gubareva LV, Katz JM, Villanueva JM, Donis RO, Cox NJ (2014) Use of highly pathogenic avian influenza A(H5N1) gain-of-function studies for molecular-based surveillance and pandemic preparedness. MBio 5(6):e02431. https://doi.org/10. 1128/mBio.02431-14
54. Lipsitch M (2016) Comment on “gain-offunction research and the relevance to clinical practice”. J Infect Dis 214(8):1284–1285. https://doi.org/10.1093/infdis/jiw348
55. Jimenez-Alberto A, Alvarado-Facundo E, Ribas-Aparicio RM, Castelan-Vega JA (2013) Analysis of adaptation mutants in the hemagglutinin of the influenza A(H1N1)pdm09 virus. PLoS One 8(7):e70005. https://doi. org/10.1371/journal.pone.0070005
56. Larsson P, Kasson PM (2013) Lipid tail protrusion in simulations predicts fusogenic activity of influenza fusion peptide mutants and conformational models. PLoS Comput Biol 9(3): e1002950. https://doi.org/10.1371/journal. pcbi.1002950
57. Ripoll DR, Khavrutskii IV, Chaudhury S, Liu J, Kuschner RA, Wallqvist A, Reifman J (2012) Quantitative predictions of binding free energy changes in drug-resistant influenza neuraminidase. PLoS Comput Biol 8(8):e1002665. https://doi.org/10.1371/journal.pcbi. 1002665
58. Shelton H, Roberts KL, Molesti E, Temperton N, Barclay WS (2013) Mutations in haemagglutinin that affect receptor binding and pH stability increase replication of a PR8 influenza virus with H5 HA in the upper respiratory tract of ferrets and may contribute to transmissibility. J Gen Virol 94 (Pt 6):1220–1229. https://doi.org/10. 1099/vir.0.050526-0
59. Shelton H, Ayora-Talavera G, Ren J, Loureiro S, Pickles RJ, Barclay WS, Jones IM (2011) Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J Virol 85(4):1875–1880. https://doi.org/10. 1128/JVI.01822-10
60. Gong LI, Suchard MA, Bloom JD (2013) Stability-mediated epistasis constrains the evolution of an influenza protein. eLife 2:e00631. https://doi.org/10.7554/eLife.00631
61. Cauldwell AV, Moncorge O, Barclay WS (2013) Unstable polymerase-nucleoprotein interaction is not responsible for avian influenza virus polymerase restriction in human cells. J Virol 87(2):1278–1284. https://doi. org/10.1128/JVI.02597-12
62. Tamuri AU, Dos Reis M, Hay AJ, Goldstein RA (2009) Identifying changes in selective constraints: host shifts in influenza. PLoS Comput Biol 5(11):e1000564. https://doi. org/10.1371/journal.pcbi.1000564
63. dos Reis M, Tamuri AU, Hay AJ, Goldstein RA (2011) Charting the host adaptation of influenza viruses. Mol Biol Evol 28(6):1755–1767. https://doi.org/10.1093/molbev/msq317
64. Jonges M, Meijer A, Fouchier RA, Koch G, Li J, Pan JC, Chen H, Shu YL, Koopmans MP (2013) Guiding outbreak management by the use of influenza A(H7Nx) virus sequence analysis. Euro Surveill 18(16):20460
65. Wong EH, Smith DK, Rabadan R, Peiris M, Poon LL (2010) Codon usage bias and the evolution of influenza A viruses. Codon usage biases of influenza virus. BMC Evol Biol 10:253. https://doi.org/10.1186/1471- 2148-10-253
66. Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H (2013) H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341 (6144):410–414. https://doi.org/10.1126/ science.1240532
67. Gustin KM, Katz JM, Tumpey TM, Maines TR (2013) Comparison of the levels of infectious virus in respirable aerosols exhaled by ferrets infected with influenza viruses exhibiting diverse transmissibility phenotypes. J Virol 87 (14):7864–7873. https://doi.org/10.1128/ JVI.00719-13
68. Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, Garcia-Sastre A, Palese P, Hai R (2011) The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the Guinea pig model. J Virol 85(21):11235–11241. https:// doi.org/10.1128/JVI.05794-11
69. Yen HL, Liang CH, Wu CY, Forrest HL, Ferguson A, Choy KT, Jones J, Wong DD, Cheung PP, Hsu CH, Li OT, Yuen KM, Chan RW, Poon LL, Chan MC, Nicholls JM, Krauss S, Wong CH, Guan Y, Webster RG, Webby RJ, Peiris M (2011) Hemagglutininneuraminidase balance confers respiratorydroplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A 108(34):14264–14269. https://doi.org/10.1073/pnas.1111000108
70. Krammer F, Palese P (2013) Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3(5):521–530. https:// doi.org/10.1016/j.coviro.2013.07.007
71. Wei CJ, Yassine HM, McTamney PM, Gall JG, Whittle JR, Boyington JC, Nabel GJ (2012) Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Sci Transl Med 4(147):147ra114. https://doi.org/10.1126/scitranslmed. 3004273
72. Schmitz N, Beerli RR, Bauer M, Jegerlehner A, Dietmeier K, Maudrich M, Pumpens P, Saudan P, Bachmann MF (2012) Universal vaccine against influenza virus: linking TLR signaling to anti-viral protection. Eur J Immunol 42(4):863–869. https://doi.org/10. 1002/eji.201041225
73. Hughes B, Hayden F, Perikov Y, Hombach J, Tam JS (2012) Report of the 5th meeting on influenza vaccines that induce broad spectrum and long-lasting immune responses, World Health Organization, Geneva, 16–- 17 November 2011. Vaccine 30 (47):6612–6622. https://doi.org/10.1016/j. vaccine.2012.08.073
74. Doyle TM, Jaentschke B, Van Domselaar G, Hashem AM, Farnsworth A, Forbes NE, Li C, Wang J, He R, Brown EG, Li X (2013) The universal epitope of influenza A viral neuraminidase fundamentally contributes to enzyme activity and viral replication. J Biol Chem 288(25):18283–18289. https://doi. org/10.1074/jbc.M113.468884
75. Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, Digard P, Anttila V, Baillie JK, Walsh TS, Hume DA, Palotie A, Xue Y, Colonna V, Tyler-Smith C, Dunning J, Gordon SB, Smyth RL, Openshaw PJ, Dougan G, Brass AL, Kellam P (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484(7395):519–523. https:// doi.org/10.1038/nature10921
76. Dormitzer PR, Suphaphiphat P, Gibson DG, Wentworth DE, Stockwell TB, Algire MA, Alperovich N, Barro M, Brown DM, Craig S, Dattilo BM, Denisova EA, De Souza I, Eickmann M, Dugan VG, Ferrari A, Gomila RC, Han L, Judge C, Mane S, Matrosovich M, Merryman C, Palladino G, Palmer GA, Spencer T, Strecker T, Trusheim H, Uhlendorff J, Wen Y, Yee AC, Zaveri J, Zhou B, Becker S, Donabedian A, Mason PW, Glass JI, Rappuoli R, Venter JC (2013) Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci Transl Med 5(185):185ra168. https://doi. org/10.1126/scitranslmed.3006368
______________
Notes:
Yohei Yamauchi (ed.), Influenza Virus: Methods and Protocols, Methods in Molecular Biology, vol. 1836, https://doi.org/10.1007/978-1-4939-8678-1_29, © Springer Science+Business Media, LLC, part of Springer Nature 2018
- admin
- Site Admin
- Posts: 36135
- Joined: Thu Aug 01, 2013 5:21 am
Re: U.S. government gave $3.7 million grant to Wuhan lab at
Why US outsourced bat virus research to Wuhan: US-funded $3.7 million project approved by Trump's Covid-19 guru Dr Anthony Fauci in 2015 after US ban imposed on 'monster-germ' research
by Christina Lin
AsiaTimes
April 22, 2020
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT

The US funded research into bat coronaviruses in a lab in Wuhan, China, that is now under scrutiny for possibly being behind the Covid-19 pandemic. Photo: Facebook
The US National Institutes of Health (NIH) funded bat-coronavirus research in the Wuhan Institute of Virology in China to the tune of US $3.7 million, a recent article in the British newspaper Daily Mail revealed.
Back in October 2014, the US government had placed a federal moratorium on gain-of-function (GOF) research -– altering natural pathogens to make them more deadly and infectious -– as a result of rising fears about a possible pandemic caused by an accidental or deliberate release of these genetically engineered monster germs.
This was in part due to lab accidents at the US Centers for Disease Control and Prevention (CDC) in July 2014 that raised questions about biosafety at US high-containment labs.
At that time, the CDC had closed two labs and halted some biological shipments in the wake of several incidents in which highly pathogenic microbes were mishandled by US government laboratories: an accidental shipment of live anthrax, the discovery of forgotten live smallpox samples and a newly revealed incident in which a dangerous influenza strain was accidentally shipped from the CDC to another lab.
A CDC internal report described how scientists failed to follow proper procedures to ensure samples were inactivated before they left the lab, and also found “multiple other problems” with operating procedures in the anthrax lab.
As such in October 2014, because of public health concerns, the US government banned all federal funding on efforts to weaponize three viruses –- influenza, Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS).

Director of the National Institute of Allergy and Infectious Diseases Anthony Fauci speaks during the daily briefing on the novel coronavirus, Covid-19, at the White House on March 24, 2020, in Washington, DC. Photo: AFP
In the face of a moratorium in the US, Dr Anthony Fauci –- the director of the National Institute of Allergy and Infectious Diseases (NIAID) and currently the leading doctor in the US Coronavirus Task Force –- outsourced in 2015 the GOF research to China’s Wuhan lab and licensed the lab to continue receiving US government funding.
The Wuhan lab is now at the center of scrutiny for possibly releasing the SARS-CoV-2 coronavirus and causing the global Covid-19 pandemic.
It is understandable that the Chinese lab likely struggled with safety issues given the fact U.S. labs share similar problems, and indeed in January 2018 the US Embassy in Beijing sent cables warning about the safety of the Wuhan lab and asked for help.
Additionally, the embassy warned that researchers “showed that various SARS-like coronaviruses can interact with ACE2, the human receptor identified for SARS-coronavirus,” meaning bat coronaviruses can be transmitted to humans to cause SARS-like diseases.
Now, the US is up in arms to hold China accountable for the global coronavirus pandemic, filing class-action lawsuits domestically, as well as building a coalition with allies internationally.
Lawsuits have been filed within the US and the International Criminal Court alleging that China used the virus as a bioweapon, and other suits are under way at the International Court of Justice. Republican lawmakers such as Senator Tom Cotton and Representative Dan Crenshaw have also introduced legislation that would allow Americans to sue China in federal court over the deaths and economic damage wrought by the virus.
US spy agencies are also investigating whether the virus originated in the Wuhan lab, and seeking evidence that is needed to support the bio-WMD theory promoted by Republican lawmakers.
If evidence is found that Covid-19 is a biological weapon, some pundits such as Fox News host Lou Dobbs have called for the US to declare war on China.
Nonetheless, it is unclear what the legal ramifications would be if the virus was indeed leaked from a Chinese lab, but as a result of a research project that was outsourced and funded by the US government.
Also, if there was a government ban in 2014 on federal funding being used for GOF research, what are the federal compliance and ethical issues surrounding the fact that the NIH still gave federal funding instead of private funding to the Wuhan lab to continue the experiments?

Hazard suits at the high-security National Biosafety Laboratory in Wuhan. Photo: Wuhan Virology Institute
Moreover, could some strains of the coronavirus have originated in U.S. labs, given the fact the US government lifted the ban in December 2017 on GOF research without resolving lab-safety issues?
For now, President Donald Trump’s administration is investigating the $3.7 million in taxpayer money that went to the Wuhan lab, while Republican Representative Matt Gaetz called for an immediate end to NIH funding of Chinese research. Since the federal ban on GOF research has been lifted, US labs can continue creating these monster germs domestically and would no longer need to outsource to China.
Nonetheless, there still needs to be better oversight on the dangerous experiments and regulations over biosecurity of labs.
Currently, the National Science Advisory Board for Biosecurity (NSABB) – a US government interagency panel that advises the NIH’s parent, the US Department of Health and Human Services (HHS) – conducts risk assessment of GOF experiments that pose a significant threat to public health.
The NSABB has given the HHS a framework to assess proposed research that would create pathogens with pandemic potential, such as research on genetically altering a virus to infect more species, or recreating a pathogen that has been eradicated in the wild, such as smallpox.
However, vaccine development and epidemiological surveillance do not automatically trigger an HHS review. In the postmortem of the Covid-19 pandemic, this is likely a dangerous loophole that could be exploited with no oversight, and should probably be brought under HHS review in order to protect public health better in the future.
by Christina Lin
AsiaTimes
April 22, 2020
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
YOU ARE REQUIRED TO READ THE COPYRIGHT NOTICE AT THIS LINK BEFORE YOU READ THE FOLLOWING WORK, THAT IS AVAILABLE SOLELY FOR PRIVATE STUDY, SCHOLARSHIP OR RESEARCH PURSUANT TO 17 U.S.C. SECTION 107 AND 108. IN THE EVENT THAT THE LIBRARY DETERMINES THAT UNLAWFUL COPYING OF THIS WORK HAS OCCURRED, THE LIBRARY HAS THE RIGHT TO BLOCK THE I.P. ADDRESS AT WHICH THE UNLAWFUL COPYING APPEARED TO HAVE OCCURRED. THANK YOU FOR RESPECTING THE RIGHTS OF COPYRIGHT OWNERS.

The US funded research into bat coronaviruses in a lab in Wuhan, China, that is now under scrutiny for possibly being behind the Covid-19 pandemic. Photo: Facebook
The US National Institutes of Health (NIH) funded bat-coronavirus research in the Wuhan Institute of Virology in China to the tune of US $3.7 million, a recent article in the British newspaper Daily Mail revealed.
Back in October 2014, the US government had placed a federal moratorium on gain-of-function (GOF) research -– altering natural pathogens to make them more deadly and infectious -– as a result of rising fears about a possible pandemic caused by an accidental or deliberate release of these genetically engineered monster germs.
This was in part due to lab accidents at the US Centers for Disease Control and Prevention (CDC) in July 2014 that raised questions about biosafety at US high-containment labs.
At that time, the CDC had closed two labs and halted some biological shipments in the wake of several incidents in which highly pathogenic microbes were mishandled by US government laboratories: an accidental shipment of live anthrax, the discovery of forgotten live smallpox samples and a newly revealed incident in which a dangerous influenza strain was accidentally shipped from the CDC to another lab.
A CDC internal report described how scientists failed to follow proper procedures to ensure samples were inactivated before they left the lab, and also found “multiple other problems” with operating procedures in the anthrax lab.
As such in October 2014, because of public health concerns, the US government banned all federal funding on efforts to weaponize three viruses –- influenza, Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS).

Director of the National Institute of Allergy and Infectious Diseases Anthony Fauci speaks during the daily briefing on the novel coronavirus, Covid-19, at the White House on March 24, 2020, in Washington, DC. Photo: AFP
In the face of a moratorium in the US, Dr Anthony Fauci –- the director of the National Institute of Allergy and Infectious Diseases (NIAID) and currently the leading doctor in the US Coronavirus Task Force –- outsourced in 2015 the GOF research to China’s Wuhan lab and licensed the lab to continue receiving US government funding.
The Wuhan lab is now at the center of scrutiny for possibly releasing the SARS-CoV-2 coronavirus and causing the global Covid-19 pandemic.
It is understandable that the Chinese lab likely struggled with safety issues given the fact U.S. labs share similar problems, and indeed in January 2018 the US Embassy in Beijing sent cables warning about the safety of the Wuhan lab and asked for help.
Additionally, the embassy warned that researchers “showed that various SARS-like coronaviruses can interact with ACE2, the human receptor identified for SARS-coronavirus,” meaning bat coronaviruses can be transmitted to humans to cause SARS-like diseases.
Now, the US is up in arms to hold China accountable for the global coronavirus pandemic, filing class-action lawsuits domestically, as well as building a coalition with allies internationally.
Lawsuits have been filed within the US and the International Criminal Court alleging that China used the virus as a bioweapon, and other suits are under way at the International Court of Justice. Republican lawmakers such as Senator Tom Cotton and Representative Dan Crenshaw have also introduced legislation that would allow Americans to sue China in federal court over the deaths and economic damage wrought by the virus.
US spy agencies are also investigating whether the virus originated in the Wuhan lab, and seeking evidence that is needed to support the bio-WMD theory promoted by Republican lawmakers.
If evidence is found that Covid-19 is a biological weapon, some pundits such as Fox News host Lou Dobbs have called for the US to declare war on China.
Nonetheless, it is unclear what the legal ramifications would be if the virus was indeed leaked from a Chinese lab, but as a result of a research project that was outsourced and funded by the US government.
Also, if there was a government ban in 2014 on federal funding being used for GOF research, what are the federal compliance and ethical issues surrounding the fact that the NIH still gave federal funding instead of private funding to the Wuhan lab to continue the experiments?

Hazard suits at the high-security National Biosafety Laboratory in Wuhan. Photo: Wuhan Virology Institute
Moreover, could some strains of the coronavirus have originated in U.S. labs, given the fact the US government lifted the ban in December 2017 on GOF research without resolving lab-safety issues?
You know Ralph, I think the public has not been in on this debate. It has been a secret debate with the NIH and other people who funded it. I think COVID-19 is a product of this. And I know Trump wants to call it the China virus. Well, the money that went into the creation and the genetic engineering of these coronaviruses in Wuhan was supported by the NIH and the USAID. So why wouldn't it be the NIH virus, or the USAID virus?
-- Interview with Andrew Kimball on the Ralph Nader Radio Show
For now, President Donald Trump’s administration is investigating the $3.7 million in taxpayer money that went to the Wuhan lab, while Republican Representative Matt Gaetz called for an immediate end to NIH funding of Chinese research. Since the federal ban on GOF research has been lifted, US labs can continue creating these monster germs domestically and would no longer need to outsource to China.
Nonetheless, there still needs to be better oversight on the dangerous experiments and regulations over biosecurity of labs.
Currently, the National Science Advisory Board for Biosecurity (NSABB) – a US government interagency panel that advises the NIH’s parent, the US Department of Health and Human Services (HHS) – conducts risk assessment of GOF experiments that pose a significant threat to public health.
The NSABB has given the HHS a framework to assess proposed research that would create pathogens with pandemic potential, such as research on genetically altering a virus to infect more species, or recreating a pathogen that has been eradicated in the wild, such as smallpox.
However, vaccine development and epidemiological surveillance do not automatically trigger an HHS review. In the postmortem of the Covid-19 pandemic, this is likely a dangerous loophole that could be exploited with no oversight, and should probably be brought under HHS review in order to protect public health better in the future.
- admin
- Site Admin
- Posts: 36135
- Joined: Thu Aug 01, 2013 5:21 am
Re: U.S. government gave $3.7 million grant to Wuhan lab at
Controversial experiments that could make bird flu more risky poised to resume
by Jocelyn Kaiser
ScienceMag.org
Feb. 8, 2019 , 8:45 PM
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
A worker at a Centers for Disease Control and Prevention laboratory harvests avian flu viruses for sharing with other laboratories in 2013. JAMES GATHANY/CDC
Controversial lab studies that modify bird flu viruses in ways that could make them more risky to humans will soon resume after being on hold for more than 4 years. ScienceInsider has learned that last year, a U.S. government review panel quietly approved experiments proposed by two labs that were previously considered so dangerous that federal officials had imposed an unusual top-down moratorium on such research.
One of the projects has already received funding from the National Institutes of Health’s (NIH’s) National Institute of Allergy and Infectious Diseases (NIAID) in Bethesda, Maryland, and will start in a few weeks; the other is awaiting funding.
The outcome may not satisfy scientists who believe certain studies that aim to make pathogens more potent or more likely to spread in mammals are so risky they should be limited or even banned. Some are upset because the government’s review will not be made public. “After a deliberative process that cost $1 million for [a consultant’s] external study and consumed countless weeks and months of time for many scientists, we are now being asked to trust a completely opaque process where the outcome is to permit the continuation of dangerous experiments,“ says Harvard University epidemiologist Marc Lipsitch.
One of the investigators leading the studies, however, says he’s happy he can resume his experiments. “We are glad the United States government weighed the risks and benefits … and developed new oversight mechanisms. We know that it does carry risks. We also believe it is important work to protect human health,” says Yoshihiro Kawaoka of the University of Wisconsin in Madison and the University of Tokyo. The other group that got the green light is led by Ron Fouchier at Erasmus University Medical Center in Rotterdam, the Netherlands.
In 2011, Fouchier and Kawaoka alarmed the world by revealing they had separately modified the deadly avian H5N1 influenza virus so that it spread between ferrets. Advocates of such gain of function (GOF) studies say they can help public health experts better understand how viruses might spread and plan for pandemics. But by enabling the bird virus to more easily spread among mammals, the experiments also raised fears that the pathogen could jump to humans. And critics of the work worried that such a souped-up virus could spark a pandemic if it escaped from a lab or was intentionally released by a bioterrorist. After extensive discussion about whether the two studies should even be published (they ultimately were) and a voluntary moratorium by the two labs, the experiments resumed in 2013 under new U.S. oversight rules.

Yoshihiro Kawaoka (left) and Ron Fouchier (right) in 2012, after their work with H5N1 bird flu virus sparked a global controversy over research that can potentially make pathogens more dangerous to humans. MARTIN ENSERINK/SCIENCE
But concerns reignited after more papers and a series of accidents at federal biocontainment labs. In October 2014, U.S. officials announced an unprecedented “pause” on funding for 18 GOF studies involving influenza or the Middle East respiratory syndrome or severe acute respiratory syndrome viruses. (About half were later allowed to continue because the work didn’t fit the definition or was deemed essential to public health.)
There followed two National Academy of Sciences workshops, recommendations from a federal advisory board, and a new U.S. policy for evaluating proposed studies involving “enhanced potential pandemic pathogens” (known as ePPPs). In December 2017, NIH lifted the funding pause and invited new GOF proposals that would be reviewed by a committee with wide-ranging expertise drawn from the Department of Health and Human Services (HHS) in Washington, D.C., and other federal agencies.
Now, the HHS committee has approved the same type of work in the Kawaoka and Fouchier labs that set off the furor 8 years ago. Last summer, the committee reviewed the projects and made recommendations about risk-benefit analyses, safety measures to avoid exposures, and communications plans, an HHS spokesperson says.
After the investigators revised their plans, the HHS committee recommended that they proceed. Kawaoka learned from NIH on 10 January that his grant has been funded. Fouchier expects the agency may hold off on making a funding decision until after a routine U.S. inspection of his lab in March.
Kawaoka’s grant is the same one on H5N1 that was paused in 2014. It includes identifying mutations in H5N1 that allow it to be transmitted by respiratory droplets in ferrets. He shared a list of reporting requirements that appear to reflect the new HHS review criteria. For example, he must immediately notify NIAID if he identifies an H5N1 strain that is both able to spread via respiratory droplets in ferrets and is highly pathogenic, or if he develops an EPPP that is resistant to antiviral drugs. Under the HHS framework, his grant now specifies reporting timelines and who he must notify at the NIAID and his university.
Fouchier’s proposed projects are part of a contract led by virologists at the Icahn School of Medicine at Mount Sinai in New York City (most of Project 5, Aim 3.1, and Project 6 in this letter). They include identifying molecular changes that make flu viruses more virulent and mutations that emerge when H5N1 is passaged through ferrets. The HHS panel did not ask that any proposed experiments be removed or modified. Suggestions included clarifying how his team will monitor workers for possible exposures and justifying the strains they plan to work with, which include H7N9 viruses, Fouchier says.
HHS cannot make the panel’s reviews public because they contain proprietary and grant competition information, says the spokesperson. But critics say that isn’t acceptable. “Details regarding the decision to approve and fund this work should be made transparent,” says Thomas Inglesby, director of Center for Health Security of the Johns Hopkins Bloomberg School of Public Health in Baltimore, Maryland. The lack of openness "is disturbing. And indefensible,” says microbiologist Richard Ebright of Rutgers University in Piscataway, New Jersey. The critics say the HHS panel should at least publicly explain why it thought the same questions could not be answered using safer alternative methods.
One researcher who has sympathized with both sides in the debate finds the safety conditions imposed on Kawaoka reassuring. “That list… makes a lot of sense,” says virologist Michael Imperiale of the University of Michigan in Ann Arbor. “At this point I’m willing to trust the system.”
by Jocelyn Kaiser
ScienceMag.org
Feb. 8, 2019 , 8:45 PM
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
YOU ARE REQUIRED TO READ THE COPYRIGHT NOTICE AT THIS LINK BEFORE YOU READ THE FOLLOWING WORK, THAT IS AVAILABLE SOLELY FOR PRIVATE STUDY, SCHOLARSHIP OR RESEARCH PURSUANT TO 17 U.S.C. SECTION 107 AND 108. IN THE EVENT THAT THE LIBRARY DETERMINES THAT UNLAWFUL COPYING OF THIS WORK HAS OCCURRED, THE LIBRARY HAS THE RIGHT TO BLOCK THE I.P. ADDRESS AT WHICH THE UNLAWFUL COPYING APPEARED TO HAVE OCCURRED. THANK YOU FOR RESPECTING THE RIGHTS OF COPYRIGHT OWNERS.

A worker at a Centers for Disease Control and Prevention laboratory harvests avian flu viruses for sharing with other laboratories in 2013. JAMES GATHANY/CDC
Controversial lab studies that modify bird flu viruses in ways that could make them more risky to humans will soon resume after being on hold for more than 4 years. ScienceInsider has learned that last year, a U.S. government review panel quietly approved experiments proposed by two labs that were previously considered so dangerous that federal officials had imposed an unusual top-down moratorium on such research.
One of the projects has already received funding from the National Institutes of Health’s (NIH’s) National Institute of Allergy and Infectious Diseases (NIAID) in Bethesda, Maryland, and will start in a few weeks; the other is awaiting funding.
The outcome may not satisfy scientists who believe certain studies that aim to make pathogens more potent or more likely to spread in mammals are so risky they should be limited or even banned. Some are upset because the government’s review will not be made public. “After a deliberative process that cost $1 million for [a consultant’s] external study and consumed countless weeks and months of time for many scientists, we are now being asked to trust a completely opaque process where the outcome is to permit the continuation of dangerous experiments,“ says Harvard University epidemiologist Marc Lipsitch.
One of the investigators leading the studies, however, says he’s happy he can resume his experiments. “We are glad the United States government weighed the risks and benefits … and developed new oversight mechanisms. We know that it does carry risks. We also believe it is important work to protect human health,” says Yoshihiro Kawaoka of the University of Wisconsin in Madison and the University of Tokyo. The other group that got the green light is led by Ron Fouchier at Erasmus University Medical Center in Rotterdam, the Netherlands.
In 2011, Fouchier and Kawaoka alarmed the world by revealing they had separately modified the deadly avian H5N1 influenza virus so that it spread between ferrets. Advocates of such gain of function (GOF) studies say they can help public health experts better understand how viruses might spread and plan for pandemics. But by enabling the bird virus to more easily spread among mammals, the experiments also raised fears that the pathogen could jump to humans. And critics of the work worried that such a souped-up virus could spark a pandemic if it escaped from a lab or was intentionally released by a bioterrorist. After extensive discussion about whether the two studies should even be published (they ultimately were) and a voluntary moratorium by the two labs, the experiments resumed in 2013 under new U.S. oversight rules.

Yoshihiro Kawaoka (left) and Ron Fouchier (right) in 2012, after their work with H5N1 bird flu virus sparked a global controversy over research that can potentially make pathogens more dangerous to humans. MARTIN ENSERINK/SCIENCE
But concerns reignited after more papers and a series of accidents at federal biocontainment labs. In October 2014, U.S. officials announced an unprecedented “pause” on funding for 18 GOF studies involving influenza or the Middle East respiratory syndrome or severe acute respiratory syndrome viruses. (About half were later allowed to continue because the work didn’t fit the definition or was deemed essential to public health.)
There followed two National Academy of Sciences workshops, recommendations from a federal advisory board, and a new U.S. policy for evaluating proposed studies involving “enhanced potential pandemic pathogens” (known as ePPPs). In December 2017, NIH lifted the funding pause and invited new GOF proposals that would be reviewed by a committee with wide-ranging expertise drawn from the Department of Health and Human Services (HHS) in Washington, D.C., and other federal agencies.
Now, the HHS committee has approved the same type of work in the Kawaoka and Fouchier labs that set off the furor 8 years ago. Last summer, the committee reviewed the projects and made recommendations about risk-benefit analyses, safety measures to avoid exposures, and communications plans, an HHS spokesperson says.
After the investigators revised their plans, the HHS committee recommended that they proceed. Kawaoka learned from NIH on 10 January that his grant has been funded. Fouchier expects the agency may hold off on making a funding decision until after a routine U.S. inspection of his lab in March.
Kawaoka’s grant is the same one on H5N1 that was paused in 2014. It includes identifying mutations in H5N1 that allow it to be transmitted by respiratory droplets in ferrets. He shared a list of reporting requirements that appear to reflect the new HHS review criteria. For example, he must immediately notify NIAID if he identifies an H5N1 strain that is both able to spread via respiratory droplets in ferrets and is highly pathogenic, or if he develops an EPPP that is resistant to antiviral drugs. Under the HHS framework, his grant now specifies reporting timelines and who he must notify at the NIAID and his university.
Fouchier’s proposed projects are part of a contract led by virologists at the Icahn School of Medicine at Mount Sinai in New York City (most of Project 5, Aim 3.1, and Project 6 in this letter). They include identifying molecular changes that make flu viruses more virulent and mutations that emerge when H5N1 is passaged through ferrets. The HHS panel did not ask that any proposed experiments be removed or modified. Suggestions included clarifying how his team will monitor workers for possible exposures and justifying the strains they plan to work with, which include H7N9 viruses, Fouchier says.
HHS cannot make the panel’s reviews public because they contain proprietary and grant competition information, says the spokesperson. But critics say that isn’t acceptable. “Details regarding the decision to approve and fund this work should be made transparent,” says Thomas Inglesby, director of Center for Health Security of the Johns Hopkins Bloomberg School of Public Health in Baltimore, Maryland. The lack of openness "is disturbing. And indefensible,” says microbiologist Richard Ebright of Rutgers University in Piscataway, New Jersey. The critics say the HHS panel should at least publicly explain why it thought the same questions could not be answered using safer alternative methods.
One researcher who has sympathized with both sides in the debate finds the safety conditions imposed on Kawaoka reassuring. “That list… makes a lot of sense,” says virologist Michael Imperiale of the University of Michigan in Ann Arbor. “At this point I’m willing to trust the system.”
- admin
- Site Admin
- Posts: 36135
- Joined: Thu Aug 01, 2013 5:21 am
Re: U.S. government gave $3.7 million grant to Wuhan lab at
Research on Highly Pathogenic H5N1 Influenza Virus: The Way Forward
by Anthony S. Fauci
DOI: 10.1128/mBio.00359-12
Copyright © 2012 Fauci
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
ABSTRACT
The voluntary moratorium on gain-of-function research related to the transmissibility of highly pathogenic H5N1 influenza virus should continue, pending the resolution of critical policy questions concerning the rationale for performing such experiments and how best to report their results. The potential benefits and risks of these experiments must be discussed and understood by multiple stakeholders, including the general public, and all decisions regarding such research must be made in a transparent manner.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Commentary
The influenza virus research community is to be commended for implementing a voluntary moratorium on “gain-of-function” experiments related to the transmissibility of highly pathogenic H5N1 influenza virus (1). As a key funder of influenza virus research, the National Institute of Allergy and Infectious Diseases, a component of the U.S. National Institutes of Health, strongly supports the continuation of this moratorium pending the resolution of critical policy issues related to the rationale for performing and reporting such experiments. We need to be certain that the fundamental purposes of this work, together with its risks and benefits, are understood by multiple stakeholders, including the general public, and that decisions are made in a transparent manner.
It is clear that the scientists who conducted the experiments that triggered this debate (2, 3), and who are among those who voluntarily signed onto the moratorium, have conducted their research properly and under the safest and most secure conditions. However, the issue that has been intensely debated is whether knowledge obtained from these experiments could inadvertently affect public health in an adverse way, even in nations multiple time zones away. Putting aside the specter of bioterrorism for the moment, consider this hypothetical scenario: an important gain-of-function experiment involving a virus with serious pandemic potential is performed in a well-regulated, world-class laboratory by experienced investigators, but the information from the experiment is then used by another scientist who does not have the same training and facilities and is not subject to the same regulations. In an unlikely but conceivable turn of events, what if that scientist becomes infected with the virus, which leads to an outbreak and ultimately triggers a pandemic? Many ask reasonable questions: given the possibility of such a scenario—however remote—should the initial experiments have been performed and/or published in the first place, and what were the processes involved in this decision?
Scientists working in this field might say—as indeed I have said—that the benefits of such experiments and the resulting knowledge outweigh the risks. It is more likely that a pandemic would occur in nature, and the need to stay ahead of such a threat is a primary reason for performing an experiment that might appear to be risky. However, we must respect that there are genuine and legitimate concerns about this type of research, both domestically and globally. We cannot expect those who have these concerns to simply take us, the scientific community, at our word that the benefits of this work outweigh the risks, nor can we ignore their calls for greater transparency, their concerns about conflicts of interest, and their efforts to engage in a dialog about whether these experiments should have been performed in the first place. Those of us in the scientific community who believe in the merits of this work have the responsibility to address these concerns thoughtfully and respectfully.
Granted, the time it takes to engage in such a dialog could potentially delay or even immobilize the conduct of certain important experiments and the publication of valuable information that could move the field forward for the good of public health. Within the research community, many have expressed concern that important research progress could come to a halt just because of the fear that someone, somewhere, might attempt to replicate these experiments sloppily. This is a valid concern. However, although influenza virus scientists are the best-informed individuals about influenza virus science, and possibly even about the true level of risk to public health, the influenza virus research community can no longer be the only player in the discussion of whether certain experiments should be done. Public opinion (domestic and global) and the judgments of independent biosafety and biosecurity experts are also critical. If we want to continue this important work, we collectively need to do a better job of articulating the scientific rationale for such experiments well before they are performed and provide discussion about the potential risk to public health, however remote. We must also not rule out the possibility that in the course of these discussions, a broad consensus might be reached that certain experiments actually should not be conducted or reported.
In this regard, as part of an interagency process, the U.S. Government is planning to augment current policy guidance related to life sciences dual-use research of concern (DURC) (4) by developing a framework for strengthening regular institutional review and oversight of certain life sciences research with high-consequence pathogens and toxins in order to identify potential DURC and mitigate risks where appropriate. This policy implementation proposal will go well beyond H5N1 influenza virus to include 15 pathogens and likely will be modified to include additional examples of DURC. It will delineate the procedures for the oversight of DURC and the responsibilities of investigators, research institutions, and the U.S. Government. Ultimately, there will also be a companion guide to help institutions identify, assess, manage, and responsibly communicate to the public about DURC.
With regard to the specific question of whether certain gain-of-function experiments related to the transmissibility of highly pathogenic H5N1 influenza virus should be conducted at all, which addresses directly the issue of the moratorium, the U.S. Government is planning to host an international workshop before the end of 2012 with important input from the National Science Advisory Board for Biosecurity and with global representation, including those with biosafety and biosecurity expertise, influenza virus and non-influenza virus scientists, and representatives of the domestic and global public. The meeting participants will consider general principles concerning the rationale for and risks and benefits of such experiments and what lines might be drawn in their conduct and/or reporting.
The game has changed for influenza virus scientists and the agencies that support them. As researchers, we must realize that we are critical players in the process of policy and decision making related to DURC, but we are not the only players. Before embarking on certain types of research, we must ask ourselves critical questions about whether there are alternative ways to answer the research questions at hand. When no reasonable alternatives exist, we must take the scientific approach to making the argument for conducting such experiments before they are performed. The voluntary moratorium on the controversial issue of gain-of-function research related to the transmissibility of highly pathogenic H5N1 influenza virus is providing us the time and space we all need to work together and get this right, and it should be continued until we do so (5).
REFERENCES
1. Fouchier RA, et al. 2012. Pause on avian flu transmission research. Science 335:400–401. doi: 10.1126/science.1219412.FREE Full TextGoogle Scholar
2. Herfst S, et al. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. Abstract/FREE Full TextGoogle Scholar
3. Imai M, et al. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428.CrossRefPubMedWeb of ScienceGoogle Scholar
4. NIH. 2012. United States Government policy for oversight of life sciences dual use research of concern. NIH, Bethesda, MD. http://oba.od.nih.gov/oba/biosecurity/P ... pdf.Google Scholar
5. Fauci A. S. 31 July 2012. The way forward in influenza research: a dialogue with the NIAID Director. Audio of presentation from the Sixth Annual Meeting of the Centers for Excellence for Influenza Research and Surveillance (CEIRS), New York, NY. http://www.niaid.nih.gov/about/director ... mp3.Google Scholar
*************************
Statement of Drs. Collins and Fauci concerning intention to lift moratorium
Nih.gov
January 24, 2013
Colorized transmission electron micrograph of Avian influenza A H5N1 viruses (seen in gold) grown in MDCK cells (seen in green).CDC/Cynthia Goldsmith
One year ago, scientists in the H5N1 influenza research community announced that they would voluntarily suspend certain “gain-of-function” experiments involving highly pathogenic avian influenza (HPAI) H5N1 viruses pending a broad international dialogue about the future direction of this research.
That dialogue — which has included experts in the life sciences, public health, biosecurity, biosafety, law, and science policy communities — has been highly productive, with numerous meetings and publications helping to clarify the most critical issues associated with this type of research.
Countries where this research is (or might be) conducted have had the opportunity to review their policies and parameters for funding, conducting, and communicating about this research. In this context, the H5N1 influenza research community has announced their intention and support for resuming their research in those countries with final guidelines in place.
The Department of Health and Human Services (HHS) has worked in a transparent and collaborative fashion to develop a framework for reviewing funding decisions regarding research that might increase mammalian transmission of HPAI H5N1 viruses by respiratory droplets. We anticipate that the final framework for HHS funding decisions regarding HPAI H5N1 gain-of-function experiments will be complete in the next several weeks. In the meantime, U.S.-funded researchers (both those working in the United States, including those in government laboratories, as well as those working overseas) have agreed not to resume these types of HPAI H5N1 gain-of-function experiments pending finalization of the HHS Framework.
Understanding how influenza viruses become human pandemic threats is vitally important to global health preparedness. We applaud the international H5N1 influenza research community for the spirit in which they instituted this extended “pause” on their work, which has provided time for thoughtful consideration of its implications.
Francis S. Collins, M.D., Ph.D.
Director, National Institutes of Health
Anthony S. Fauci, M.D.
Director, National Institute of Allergy and Infectious Diseases
National Institutes of Health
*************************************
Dr. Fauci Backed Controversial Wuhan Lab with U.S. Dollars for Risky Coronavirus Research
by Fred Guterl
Newsweek
4/28/20 AT 2:57 PM EDT
Dr. Anthony Fauci is an adviser to President Donald Trump and something of an American folk hero for his steady, calm leadership during the pandemic crisis. At least one poll shows that Americans trust Fauci more than Trump on the coronavirus pandemic—and few scientists are portrayed on TV by Brad Pitt.
But just last year, the National Institute for Allergy and Infectious Diseases, the organization led by Dr. Fauci, funded scientists at the Wuhan Institute of Virology and other institutions for work on gain-of-function research on bat coronaviruses.
In 2019, with the backing of NIAID, the National Institutes of Health committed $3.7 million over six years for research that included some gain-of-function work. The program followed another $3.7 million, 5-year project for collecting and studying bat coronaviruses, which ended in 2019, bringing the total to $7.4 million.
Many scientists have criticized gain of function research, which involves manipulating viruses in the lab to explore their potential for infecting humans, because it creates a risk of starting a pandemic from accidental release.
SARS-CoV-2, the virus now causing a global pandemic, is believed to have originated in bats. U.S. intelligence, after originally asserting that the coronavirus had occurred naturally, conceded last month that the pandemic may have originated in a leak from the Wuhan lab. (At this point most scientists say it's possible—but not likely—that the pandemic virus was engineered or manipulated.)
Dr. Fauci did not respond to Newsweek's requests for comment. NIH responded with a statement that said in part: "Most emerging human viruses come from wildlife, and these represent a significant threat to public health and biosecurity in the US and globally, as demonstrated by the SARS epidemic of 2002-03, and the current COVID-19 pandemic.... scientific research indicates that there is no evidence that suggests the virus was created in a laboratory."
The NIH research consisted of two parts. The first part began in 2014 and involved surveillance of bat coronaviruses, and had a budget of $3.7 million. The program funded Shi Zheng-Li, a virologist at the Wuhan lab, and other researchers to investigate and catalogue bat coronaviruses in the wild. This part of the project was completed in 2019.
A second phase of the project, beginning that year, included additional surveillance work but also gain-of-function research for the purpose of understanding how bat coronaviruses could mutate to attack humans. The project was run by EcoHealth Alliance, a non-profit research group, under the direction of President Peter Daszak, an expert on disease ecology. NIH canceled the project just this past Friday, April 24th, Politico reported. Daszak did not immediately respond to Newsweek requests for comment.
The project proposal states: "We will use S protein sequence data, infectious clone technology, in vitro and in vivo infection experiments and analysis of receptor binding to test the hypothesis that % divergence thresholds in S protein sequences predict spillover potential."
In layman's terms, "spillover potential" refers to the ability of a virus to jump from animals to humans, which requires that the virus be able to attach to receptors in the cells of humans. SARS-CoV-2, for instance, is adept at binding to the ACE2 receptor in human lungs and other organs.
According to Richard Ebright, an infectious disease expert at Rutgers University, the project description refers to experiments that would enhance the ability of bat coronavirus to infect human cells and laboratory animals using techniques of genetic engineering. In the wake of the pandemic, that is a noteworthy detail.
Ebright, along with many other scientists, has been a vocal opponent of gain-of-function research because of the risk it presents of creating a pandemic through accidental release from a lab.
Dr. Fauci is renowned for his work on the HIV/AIDS crisis in the 1990s. Born in Brooklyn, he graduated first in his class from Cornell University Medical College in 1966. As head of NIAID since 1984, he has served as an adviser to every U.S. president since Ronald Reagan.
A decade ago, during a controversy over gain-of-function research on bird-flu viruses, Dr. Fauci played an important role in promoting the work. He argued that the research was worth the risk it entailed because it enables scientists to make preparations, such as investigating possible anti-viral medications, that could be useful if and when a pandemic occurred.
The work in question was a type of gain-of-function research that involved taking wild viruses and passing them through live animals until they mutate into a form that could pose a pandemic threat. Scientists used it to take a virus that was poorly transmitted among humans and make it into one that was highly transmissible—a hallmark of a pandemic virus. This work was done by infecting a series of ferrets, allowing the virus to mutate until a ferret that hadn't been deliberately infected contracted the disease.
The work entailed risks that worried even seasoned researchers. More than 200 scientists called for the work to be halted. The problem, they said, is that it increased the likelihood that a pandemic would occur through a laboratory accident.
Dr. Fauci defended the work. "[D]etermining the molecular Achilles' heel of these viruses can allow scientists to identify novel antiviral drug targets that could be used to prevent infection in those at risk or to better treat those who become infected," wrote Fauci and two co-authors in the Washington Post on December 30, 2011. "Decades of experience tells us that disseminating information gained through biomedical research to legitimate scientists and health officials provides a critical foundation for generating appropriate countermeasures and, ultimately, protecting the public health."
Nevertheless, in 2014, under pressure from the Obama administration, the National of Institutes of Health instituted a moratorium on the work, suspending 21 studies.
Three years later, though—in December 2017—the NIH ended the moratorium and the second phase of the NIAID project, which included the gain-of-function research, began. The NIH established a framework for determining how the research would go forward: scientists have to get approval from a panel of experts, who would decide whether the risks were justified.
The reviews were indeed conducted—but in secret, for which the NIH has drawn criticism. In early 2019, after a reporter for Science magazine discovered that the NIH had approved two influenza research projects that used gain of function methods, scientists who oppose this kind of research excoriated the NIH in an editorial in the Washington Post.
"We have serious doubts about whether these experiments should be conducted at all," wrote Tom Inglesby of Johns Hopkins University and Marc Lipsitch of Harvard. "[W]ith deliberations kept behind closed doors, none of us will have the opportunity to understand how the government arrived at these decisions or to judge the rigor and integrity of that process."
Correction 5/5, 6:20 p.m.: The headline of this story has been corrected to reflect that the Wuhan lab received only a part of the millions of U.S. dollars allocated for virus research.
by Anthony S. Fauci
DOI: 10.1128/mBio.00359-12
Copyright © 2012 Fauci
NOTICE: THIS WORK MAY BE PROTECTED BY COPYRIGHT
YOU ARE REQUIRED TO READ THE COPYRIGHT NOTICE AT THIS LINK BEFORE YOU READ THE FOLLOWING WORK, THAT IS AVAILABLE SOLELY FOR PRIVATE STUDY, SCHOLARSHIP OR RESEARCH PURSUANT TO 17 U.S.C. SECTION 107 AND 108. IN THE EVENT THAT THE LIBRARY DETERMINES THAT UNLAWFUL COPYING OF THIS WORK HAS OCCURRED, THE LIBRARY HAS THE RIGHT TO BLOCK THE I.P. ADDRESS AT WHICH THE UNLAWFUL COPYING APPEARED TO HAVE OCCURRED. THANK YOU FOR RESPECTING THE RIGHTS OF COPYRIGHT OWNERS.
ABSTRACT
The voluntary moratorium on gain-of-function research related to the transmissibility of highly pathogenic H5N1 influenza virus should continue, pending the resolution of critical policy questions concerning the rationale for performing such experiments and how best to report their results. The potential benefits and risks of these experiments must be discussed and understood by multiple stakeholders, including the general public, and all decisions regarding such research must be made in a transparent manner.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Commentary
The influenza virus research community is to be commended for implementing a voluntary moratorium on “gain-of-function” experiments related to the transmissibility of highly pathogenic H5N1 influenza virus (1). As a key funder of influenza virus research, the National Institute of Allergy and Infectious Diseases, a component of the U.S. National Institutes of Health, strongly supports the continuation of this moratorium pending the resolution of critical policy issues related to the rationale for performing and reporting such experiments. We need to be certain that the fundamental purposes of this work, together with its risks and benefits, are understood by multiple stakeholders, including the general public, and that decisions are made in a transparent manner.
It is clear that the scientists who conducted the experiments that triggered this debate (2, 3), and who are among those who voluntarily signed onto the moratorium, have conducted their research properly and under the safest and most secure conditions. However, the issue that has been intensely debated is whether knowledge obtained from these experiments could inadvertently affect public health in an adverse way, even in nations multiple time zones away. Putting aside the specter of bioterrorism for the moment, consider this hypothetical scenario: an important gain-of-function experiment involving a virus with serious pandemic potential is performed in a well-regulated, world-class laboratory by experienced investigators, but the information from the experiment is then used by another scientist who does not have the same training and facilities and is not subject to the same regulations. In an unlikely but conceivable turn of events, what if that scientist becomes infected with the virus, which leads to an outbreak and ultimately triggers a pandemic? Many ask reasonable questions: given the possibility of such a scenario—however remote—should the initial experiments have been performed and/or published in the first place, and what were the processes involved in this decision?
Scientists working in this field might say—as indeed I have said—that the benefits of such experiments and the resulting knowledge outweigh the risks. It is more likely that a pandemic would occur in nature, and the need to stay ahead of such a threat is a primary reason for performing an experiment that might appear to be risky. However, we must respect that there are genuine and legitimate concerns about this type of research, both domestically and globally. We cannot expect those who have these concerns to simply take us, the scientific community, at our word that the benefits of this work outweigh the risks, nor can we ignore their calls for greater transparency, their concerns about conflicts of interest, and their efforts to engage in a dialog about whether these experiments should have been performed in the first place. Those of us in the scientific community who believe in the merits of this work have the responsibility to address these concerns thoughtfully and respectfully.
Granted, the time it takes to engage in such a dialog could potentially delay or even immobilize the conduct of certain important experiments and the publication of valuable information that could move the field forward for the good of public health. Within the research community, many have expressed concern that important research progress could come to a halt just because of the fear that someone, somewhere, might attempt to replicate these experiments sloppily. This is a valid concern. However, although influenza virus scientists are the best-informed individuals about influenza virus science, and possibly even about the true level of risk to public health, the influenza virus research community can no longer be the only player in the discussion of whether certain experiments should be done. Public opinion (domestic and global) and the judgments of independent biosafety and biosecurity experts are also critical. If we want to continue this important work, we collectively need to do a better job of articulating the scientific rationale for such experiments well before they are performed and provide discussion about the potential risk to public health, however remote. We must also not rule out the possibility that in the course of these discussions, a broad consensus might be reached that certain experiments actually should not be conducted or reported.
In this regard, as part of an interagency process, the U.S. Government is planning to augment current policy guidance related to life sciences dual-use research of concern (DURC) (4) by developing a framework for strengthening regular institutional review and oversight of certain life sciences research with high-consequence pathogens and toxins in order to identify potential DURC and mitigate risks where appropriate. This policy implementation proposal will go well beyond H5N1 influenza virus to include 15 pathogens and likely will be modified to include additional examples of DURC. It will delineate the procedures for the oversight of DURC and the responsibilities of investigators, research institutions, and the U.S. Government. Ultimately, there will also be a companion guide to help institutions identify, assess, manage, and responsibly communicate to the public about DURC.
With regard to the specific question of whether certain gain-of-function experiments related to the transmissibility of highly pathogenic H5N1 influenza virus should be conducted at all, which addresses directly the issue of the moratorium, the U.S. Government is planning to host an international workshop before the end of 2012 with important input from the National Science Advisory Board for Biosecurity and with global representation, including those with biosafety and biosecurity expertise, influenza virus and non-influenza virus scientists, and representatives of the domestic and global public. The meeting participants will consider general principles concerning the rationale for and risks and benefits of such experiments and what lines might be drawn in their conduct and/or reporting.
The game has changed for influenza virus scientists and the agencies that support them. As researchers, we must realize that we are critical players in the process of policy and decision making related to DURC, but we are not the only players. Before embarking on certain types of research, we must ask ourselves critical questions about whether there are alternative ways to answer the research questions at hand. When no reasonable alternatives exist, we must take the scientific approach to making the argument for conducting such experiments before they are performed. The voluntary moratorium on the controversial issue of gain-of-function research related to the transmissibility of highly pathogenic H5N1 influenza virus is providing us the time and space we all need to work together and get this right, and it should be continued until we do so (5).
REFERENCES
1. Fouchier RA, et al. 2012. Pause on avian flu transmission research. Science 335:400–401. doi: 10.1126/science.1219412.FREE Full TextGoogle Scholar
2. Herfst S, et al. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. Abstract/FREE Full TextGoogle Scholar
3. Imai M, et al. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428.CrossRefPubMedWeb of ScienceGoogle Scholar
4. NIH. 2012. United States Government policy for oversight of life sciences dual use research of concern. NIH, Bethesda, MD. http://oba.od.nih.gov/oba/biosecurity/P ... pdf.Google Scholar
5. Fauci A. S. 31 July 2012. The way forward in influenza research: a dialogue with the NIAID Director. Audio of presentation from the Sixth Annual Meeting of the Centers for Excellence for Influenza Research and Surveillance (CEIRS), New York, NY. http://www.niaid.nih.gov/about/director ... mp3.Google Scholar
*************************
Statement of Drs. Collins and Fauci concerning intention to lift moratorium
Nih.gov
January 24, 2013

Colorized transmission electron micrograph of Avian influenza A H5N1 viruses (seen in gold) grown in MDCK cells (seen in green).CDC/Cynthia Goldsmith
One year ago, scientists in the H5N1 influenza research community announced that they would voluntarily suspend certain “gain-of-function” experiments involving highly pathogenic avian influenza (HPAI) H5N1 viruses pending a broad international dialogue about the future direction of this research.
That dialogue — which has included experts in the life sciences, public health, biosecurity, biosafety, law, and science policy communities — has been highly productive, with numerous meetings and publications helping to clarify the most critical issues associated with this type of research.
Countries where this research is (or might be) conducted have had the opportunity to review their policies and parameters for funding, conducting, and communicating about this research. In this context, the H5N1 influenza research community has announced their intention and support for resuming their research in those countries with final guidelines in place.
The Department of Health and Human Services (HHS) has worked in a transparent and collaborative fashion to develop a framework for reviewing funding decisions regarding research that might increase mammalian transmission of HPAI H5N1 viruses by respiratory droplets. We anticipate that the final framework for HHS funding decisions regarding HPAI H5N1 gain-of-function experiments will be complete in the next several weeks. In the meantime, U.S.-funded researchers (both those working in the United States, including those in government laboratories, as well as those working overseas) have agreed not to resume these types of HPAI H5N1 gain-of-function experiments pending finalization of the HHS Framework.
Understanding how influenza viruses become human pandemic threats is vitally important to global health preparedness. We applaud the international H5N1 influenza research community for the spirit in which they instituted this extended “pause” on their work, which has provided time for thoughtful consideration of its implications.
Francis S. Collins, M.D., Ph.D.
Director, National Institutes of Health
Anthony S. Fauci, M.D.
Director, National Institute of Allergy and Infectious Diseases
National Institutes of Health
*************************************
Dr. Fauci Backed Controversial Wuhan Lab with U.S. Dollars for Risky Coronavirus Research
by Fred Guterl
Newsweek
4/28/20 AT 2:57 PM EDT
Dr. Anthony Fauci is an adviser to President Donald Trump and something of an American folk hero for his steady, calm leadership during the pandemic crisis. At least one poll shows that Americans trust Fauci more than Trump on the coronavirus pandemic—and few scientists are portrayed on TV by Brad Pitt.
But just last year, the National Institute for Allergy and Infectious Diseases, the organization led by Dr. Fauci, funded scientists at the Wuhan Institute of Virology and other institutions for work on gain-of-function research on bat coronaviruses.
In 2019, with the backing of NIAID, the National Institutes of Health committed $3.7 million over six years for research that included some gain-of-function work. The program followed another $3.7 million, 5-year project for collecting and studying bat coronaviruses, which ended in 2019, bringing the total to $7.4 million.
Many scientists have criticized gain of function research, which involves manipulating viruses in the lab to explore their potential for infecting humans, because it creates a risk of starting a pandemic from accidental release.
SARS-CoV-2, the virus now causing a global pandemic, is believed to have originated in bats. U.S. intelligence, after originally asserting that the coronavirus had occurred naturally, conceded last month that the pandemic may have originated in a leak from the Wuhan lab. (At this point most scientists say it's possible—but not likely—that the pandemic virus was engineered or manipulated.)
Dr. Fauci did not respond to Newsweek's requests for comment. NIH responded with a statement that said in part: "Most emerging human viruses come from wildlife, and these represent a significant threat to public health and biosecurity in the US and globally, as demonstrated by the SARS epidemic of 2002-03, and the current COVID-19 pandemic.... scientific research indicates that there is no evidence that suggests the virus was created in a laboratory."
The NIH research consisted of two parts. The first part began in 2014 and involved surveillance of bat coronaviruses, and had a budget of $3.7 million. The program funded Shi Zheng-Li, a virologist at the Wuhan lab, and other researchers to investigate and catalogue bat coronaviruses in the wild. This part of the project was completed in 2019.
A second phase of the project, beginning that year, included additional surveillance work but also gain-of-function research for the purpose of understanding how bat coronaviruses could mutate to attack humans. The project was run by EcoHealth Alliance, a non-profit research group, under the direction of President Peter Daszak, an expert on disease ecology. NIH canceled the project just this past Friday, April 24th, Politico reported. Daszak did not immediately respond to Newsweek requests for comment.
The project proposal states: "We will use S protein sequence data, infectious clone technology, in vitro and in vivo infection experiments and analysis of receptor binding to test the hypothesis that % divergence thresholds in S protein sequences predict spillover potential."
In layman's terms, "spillover potential" refers to the ability of a virus to jump from animals to humans, which requires that the virus be able to attach to receptors in the cells of humans. SARS-CoV-2, for instance, is adept at binding to the ACE2 receptor in human lungs and other organs.
According to Richard Ebright, an infectious disease expert at Rutgers University, the project description refers to experiments that would enhance the ability of bat coronavirus to infect human cells and laboratory animals using techniques of genetic engineering. In the wake of the pandemic, that is a noteworthy detail.
Ebright, along with many other scientists, has been a vocal opponent of gain-of-function research because of the risk it presents of creating a pandemic through accidental release from a lab.
Dr. Fauci is renowned for his work on the HIV/AIDS crisis in the 1990s. Born in Brooklyn, he graduated first in his class from Cornell University Medical College in 1966. As head of NIAID since 1984, he has served as an adviser to every U.S. president since Ronald Reagan.
A decade ago, during a controversy over gain-of-function research on bird-flu viruses, Dr. Fauci played an important role in promoting the work. He argued that the research was worth the risk it entailed because it enables scientists to make preparations, such as investigating possible anti-viral medications, that could be useful if and when a pandemic occurred.
The work in question was a type of gain-of-function research that involved taking wild viruses and passing them through live animals until they mutate into a form that could pose a pandemic threat. Scientists used it to take a virus that was poorly transmitted among humans and make it into one that was highly transmissible—a hallmark of a pandemic virus. This work was done by infecting a series of ferrets, allowing the virus to mutate until a ferret that hadn't been deliberately infected contracted the disease.
The work entailed risks that worried even seasoned researchers. More than 200 scientists called for the work to be halted. The problem, they said, is that it increased the likelihood that a pandemic would occur through a laboratory accident.
H5N1 is a type of influenza virus that causes a highly infectious, severe respiratory disease in birds called avian influenza (or "bird flu"). Human cases of H5N1 avian influenza occur occasionally, but it is difficult to transmit the infection from person to person. When people do become infected, the mortality rate is about 60%.
-- FAQs: H5N1 influenza, by World Health Organization
Dr. Fauci defended the work. "[D]etermining the molecular Achilles' heel of these viruses can allow scientists to identify novel antiviral drug targets that could be used to prevent infection in those at risk or to better treat those who become infected," wrote Fauci and two co-authors in the Washington Post on December 30, 2011. "Decades of experience tells us that disseminating information gained through biomedical research to legitimate scientists and health officials provides a critical foundation for generating appropriate countermeasures and, ultimately, protecting the public health."
Nevertheless, in 2014, under pressure from the Obama administration, the National of Institutes of Health instituted a moratorium on the work, suspending 21 studies.
Three years later, though—in December 2017—the NIH ended the moratorium and the second phase of the NIAID project, which included the gain-of-function research, began. The NIH established a framework for determining how the research would go forward: scientists have to get approval from a panel of experts, who would decide whether the risks were justified.
The reviews were indeed conducted—but in secret, for which the NIH has drawn criticism. In early 2019, after a reporter for Science magazine discovered that the NIH had approved two influenza research projects that used gain of function methods, scientists who oppose this kind of research excoriated the NIH in an editorial in the Washington Post.
"We have serious doubts about whether these experiments should be conducted at all," wrote Tom Inglesby of Johns Hopkins University and Marc Lipsitch of Harvard. "[W]ith deliberations kept behind closed doors, none of us will have the opportunity to understand how the government arrived at these decisions or to judge the rigor and integrity of that process."
Correction 5/5, 6:20 p.m.: The headline of this story has been corrected to reflect that the Wuhan lab received only a part of the millions of U.S. dollars allocated for virus research.
- admin
- Site Admin
- Posts: 36135
- Joined: Thu Aug 01, 2013 5:21 am
Who is online
Users browsing this forum: No registered users and 4 guests

